Publications
For an updated list please visit Google Scholar. * denotes equal contribution.
Conference and Journal Publications
2024
- NeurIPS 2024
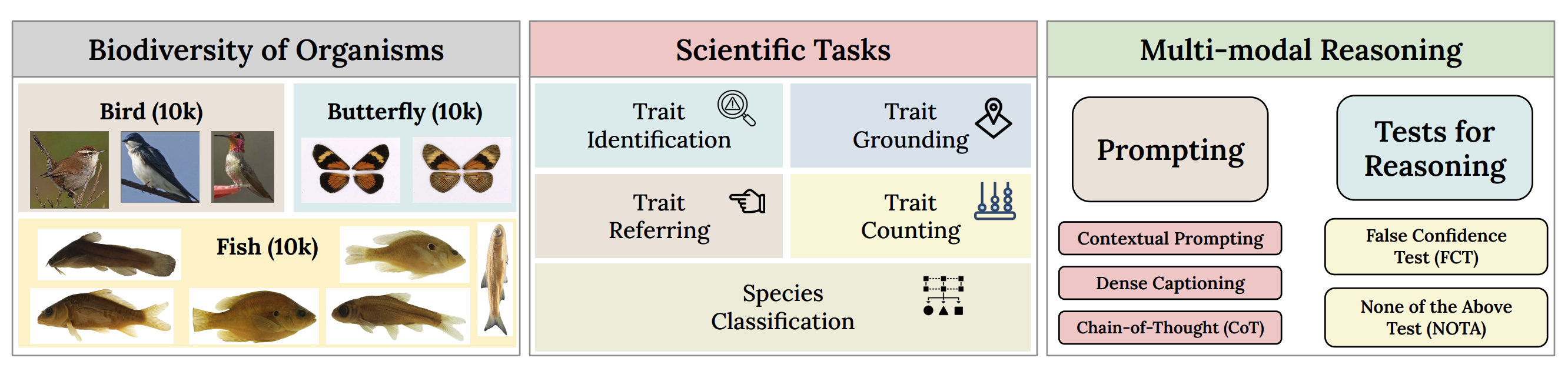 VLM4Bio: A Benchmark Dataset to Evaluate Pretrained Vision-Language Models for Trait Discovery from Biological ImagesM Maruf, Arka Daw, Kazi Sajeed Mehrab, and 8 more authorsin proceedings of Neural Information Processing Systems (NeurIPS), 2024
VLM4Bio: A Benchmark Dataset to Evaluate Pretrained Vision-Language Models for Trait Discovery from Biological ImagesM Maruf, Arka Daw, Kazi Sajeed Mehrab, and 8 more authorsin proceedings of Neural Information Processing Systems (NeurIPS), 2024Images are increasingly becoming the currency for documenting biodiversity on the planet, providing novel opportunities for accelerating scientific discoveries in the field of organismal biology, especially with the advent of large vision-language models (VLMs). We ask if pre-trained VLMs can aid scientists in answering a range of biologically relevant questions without any additional fine-tuning. In this paper, we evaluate the effectiveness of 12 state-of-the-art (SOTA) VLMs in the field of organismal biology using a novel dataset, VLM4Bio, consisting of 469K question-answer pairs involving 30K images from three groups of organisms: fishes, birds, and butterflies, covering five biologically relevant tasks. We also explore the effects of applying prompting techniques and tests for reasoning hallucination on the performance of VLMs, shedding new light on the capabilities of current SOTA VLMs in answering biologically relevant questions using images. The code and datasets for running all the analyses reported in this paper can be found at this https URL.
@article{maruf2024vlm4bio, title = {VLM4Bio: A Benchmark Dataset to Evaluate Pretrained Vision-Language Models for Trait Discovery from Biological Images}, author = {Maruf, M and Daw, Arka and Mehrab, Kazi Sajeed and Manogaran, Harish Babu and Neog, Abhilash and Sawhney, Medha and Khurana, Mridul and Balhoff, James P and Bakis, Yasin and Altintas, Bahadir and others}, journal = {in proceedings of Neural Information Processing Systems (NeurIPS)}, year = {2024}, publisher = {NeurIPS}, category = {Conference Publications} } - ECCV 2024
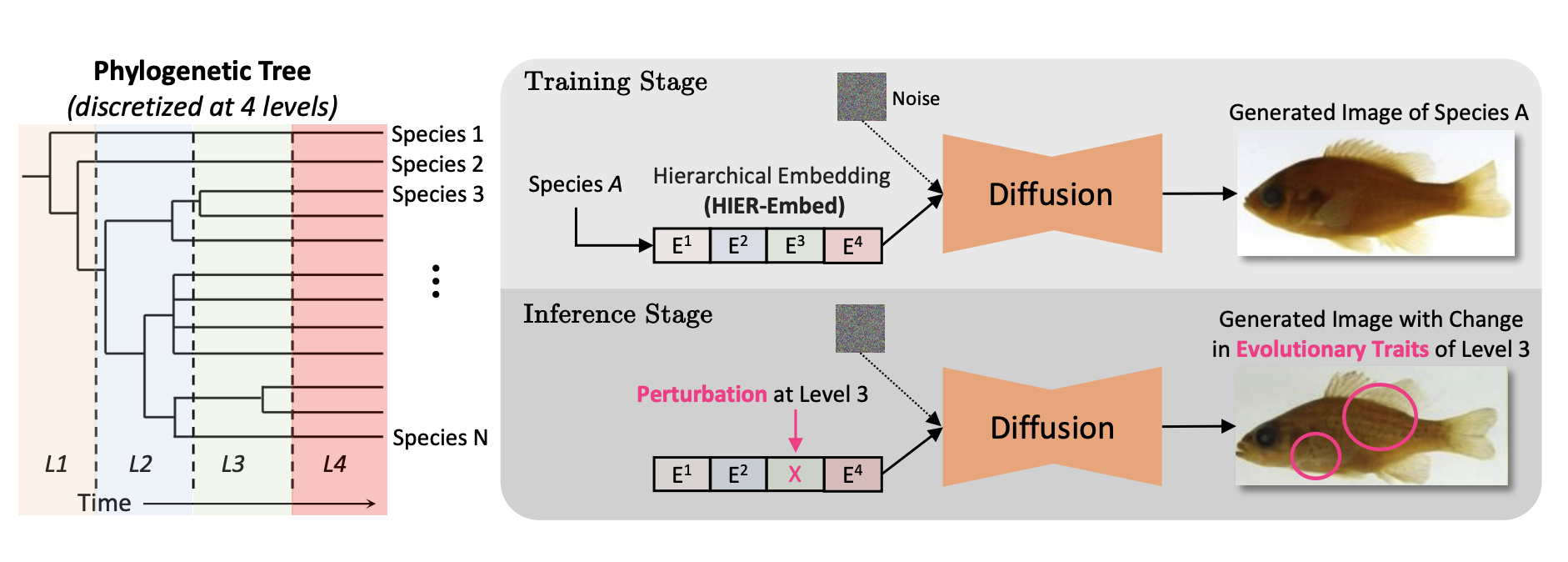 Hierarchical Conditioning of Diffusion Models Using Tree-of-Life for Studying Species EvolutionMridul Khurana, Arka Daw, M Maruf, and 8 more authorsin proceedings of European Conference on Computer Vision (ECCV), 2024
Hierarchical Conditioning of Diffusion Models Using Tree-of-Life for Studying Species EvolutionMridul Khurana, Arka Daw, M Maruf, and 8 more authorsin proceedings of European Conference on Computer Vision (ECCV), 2024A central problem in biology is to understand how organisms evolve and adapt to their environment by acquiring variations in the observable characteristics or traits of species across the tree of life. With the growing availability of large-scale image repositories in biology and recent advances in generative modeling, there is an opportunity to accelerate the discovery of evolutionary traits automatically from images. Toward this goal, we introduce Phylo-Diffusion, a novel framework for conditioning diffusion models with phylogenetic knowledge represented in the form of HIERarchical Embeddings (HIER-Embeds). We also propose two new experiments for perturbing the embedding space of Phylo-Diffusion: trait masking and trait swapping, inspired by counterpart experiments of gene knockout and gene editing/swapping. Our work represents a novel methodological advance in generative modeling to structure the embedding space of diffusion models using tree-based knowledge. Our work also opens a new chapter of research in evolutionary biology by using generative models to visualize evolutionary changes directly from images. We empirically demonstrate the usefulness of Phylo-Diffusion in capturing meaningful trait variations for fishes and birds, revealing novel insights about the biological mechanisms of their evolution.
@article{khurana2024hierarchical, title = {Hierarchical Conditioning of Diffusion Models Using Tree-of-Life for Studying Species Evolution}, author = {Khurana, Mridul and Daw, Arka and Maruf, M and Uyeda, Josef C and Dahdul, Wasila and Charpentier, Caleb and Bak{\i}{\c{s}}, Yasin and Bart Jr, Henry L and Mabee, Paula M and Lapp, Hilmar and others}, journal = {in proceedings of European Conference on Computer Vision (ECCV)}, year = {2024}, publisher = {ECCV}, category = {Conference Publications} } - AIS
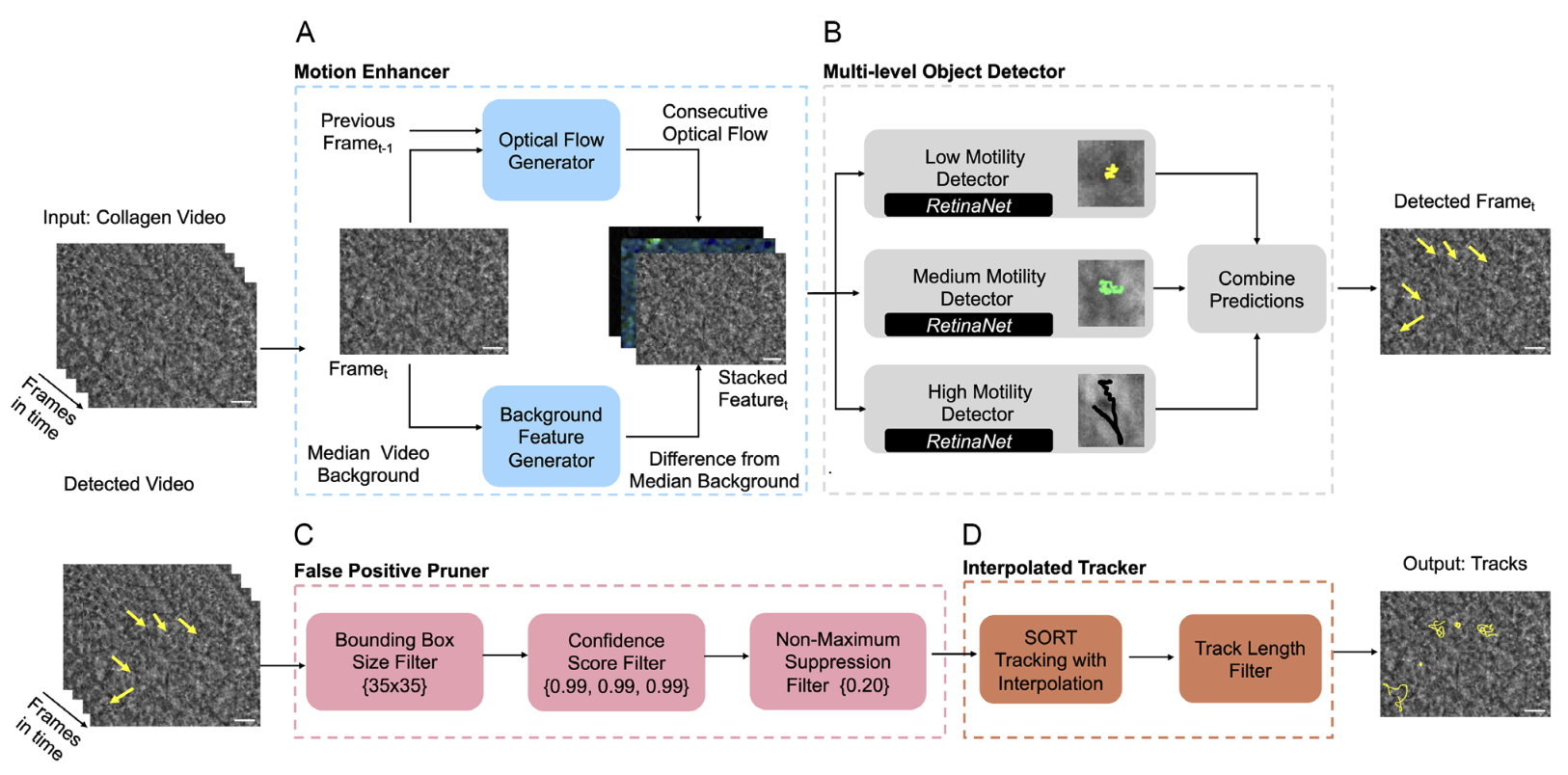 Motion Enhanced Multi-Level Tracker (MEMTrack): A Deep Learning-Based Approach to Microrobot Tracking in Dense and Low-Contrast EnvironmentsMedha Sawhney, Bhas Karmarkar, Eric J Leaman, and 3 more authorsAdvanced Intelligent Systems, 2024
Motion Enhanced Multi-Level Tracker (MEMTrack): A Deep Learning-Based Approach to Microrobot Tracking in Dense and Low-Contrast EnvironmentsMedha Sawhney, Bhas Karmarkar, Eric J Leaman, and 3 more authorsAdvanced Intelligent Systems, 2024Tracking microrobots is challenging due to their minute size and high speed. In biomedical applications, this challenge is exacerbated by the dense surrounding environments with feature sizes and shapes comparable to microrobots. Herein, Motion Enhanced Multi-level Tracker (MEMTrack) is introduced for detecting and tracking microrobots in dense and low-contrast environments. Informed by the physics of microrobot motion, synthetic motion features for deep learning-based object detection and a modified Simple Online and Real-time Tracking (SORT)algorithm with interpolation are used for tracking. MEMTrack is trained and tested using bacterial micromotors in collagen (tissue phantom), achieving precision and recall of 76% and 51%, respectively. Compared to the state-of-the-art baseline models, MEMTrack provides a minimum of 2.6-fold higher precision with a reasonably high recall. MEMTrack’s generalizability to unseen (aqueous) media and its versatility in tracking microrobots of different shapes, sizes, and motion characteristics are shown. Finally, it is shown that MEMTrack localizes objects with a root-mean-square error of less than 1.84 μm and quantifies the average speed of all tested systems with no statistically significant difference from the laboriously produced manual tracking data. MEMTrack significantly advances microrobot localization and tracking in dense and low-contrast settings and can impact fundamental and translational microrobotic research.
@article{sawhney2024motion, title = {Motion Enhanced Multi-Level Tracker (MEMTrack): A Deep Learning-Based Approach to Microrobot Tracking in Dense and Low-Contrast Environments}, author = {Sawhney, Medha and Karmarkar, Bhas and Leaman, Eric J and Daw, Arka and Karpatne, Anuj and Behkam, Bahareh}, journal = {Advanced Intelligent Systems}, volume = {6}, number = {4}, pages = {2300590}, year = {2024}, publisher = {Wiley Online Library}, category = {Conference Publications} } - Ph.D. Dissertation
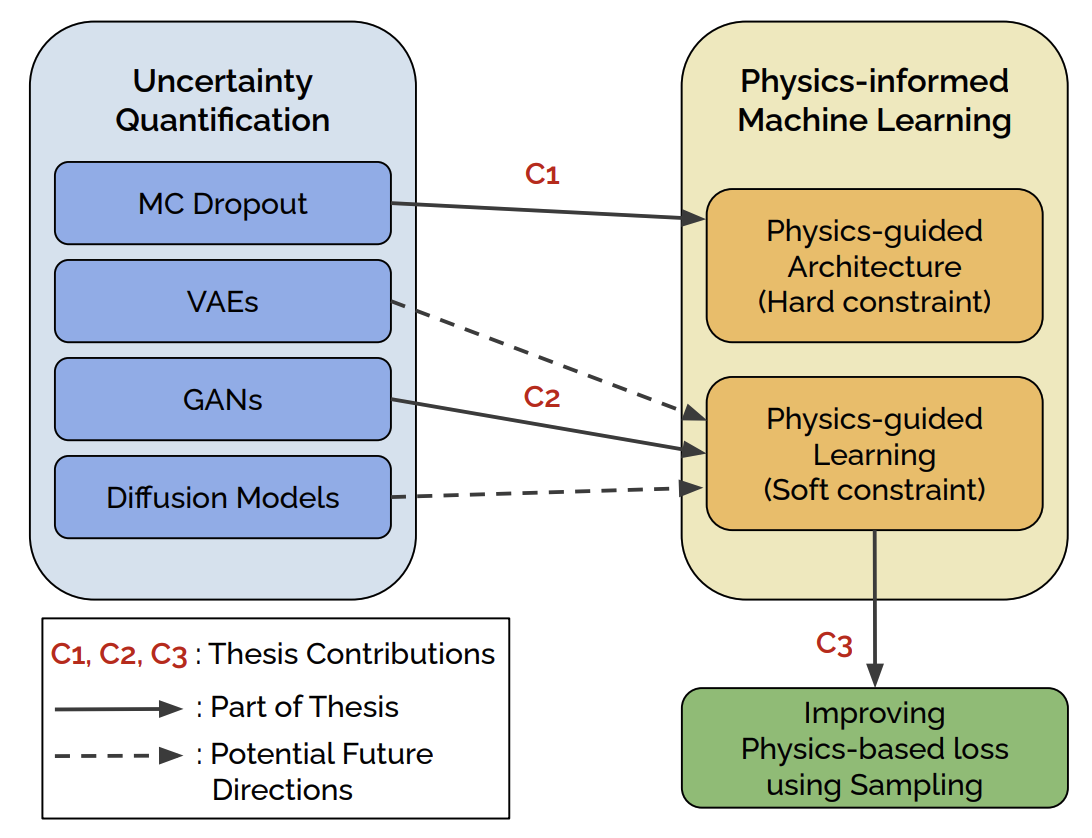 Physics-informed Machine Learning with Uncertainty QuantificationArka DawVirginia Tech, 2024
Physics-informed Machine Learning with Uncertainty QuantificationArka DawVirginia Tech, 2024Physics Informed Machine Learning (PIML) has emerged as the forefront of research in scientific machine learning with the key motivation of systematically coupling machine learning (ML) methods with prior domain knowledge often available in the form of physics supervision. Uncertainty quantification (UQ) is an important goal in many scientific use-cases, where the obtaining reliable ML model predictions and accessing the potential risks associated with them is crucial. In this thesis, we propose novel methodologies in three key areas for improving uncertainty quantification for PIML. First, we propose to explicitly infuse the physics prior in the form of monotonicity constraints through architectural modifications in neural networks for quantifying uncertainty. Second, we demonstrate a more general framework for quantifying uncertainty with PIML that is compatible with generic forms of physics supervision such as PDEs and closed form equations. Lastly, we study the limitations of physics-based loss in the context of Physics-informed Neural Networks (PINNs), and develop an efficient sampling strategy to mitigate the failure modes.
@article{daw2024physics, title = {Physics-informed Machine Learning with Uncertainty Quantification}, author = {Daw, Arka}, year = {2024}, journal = {Virginia Tech}, publisher = {Virginia Tech}, category = {Conference Publications} } - JAMES
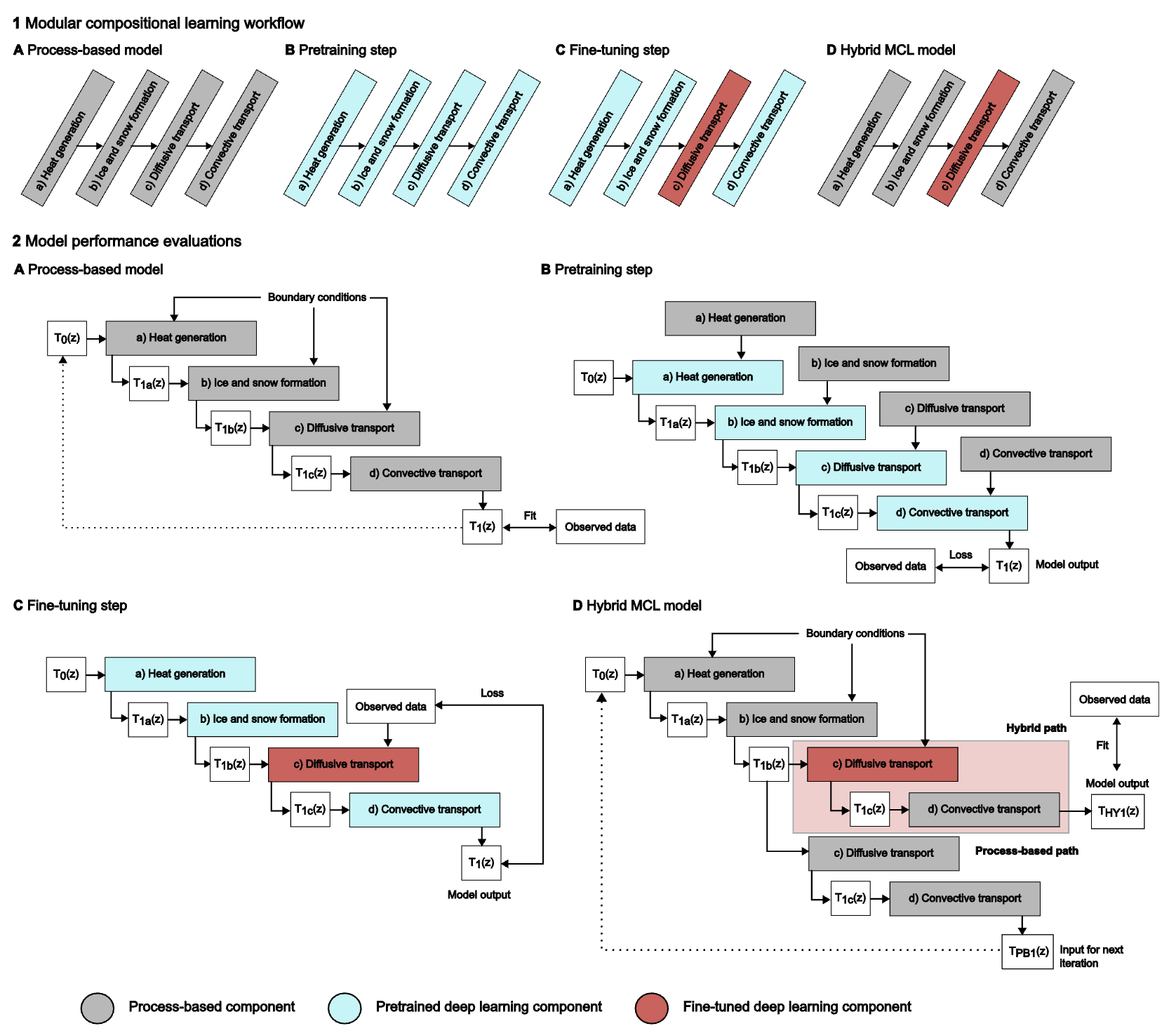 Modular Compositional Learning Improves 1D Hydrodynamic Lake Model Performance by Merging Process-Based Modeling With Deep LearningRobert Ladwig, Arka Daw, Ellen A Albright, and 6 more authorsJournal of Advances in Modeling Earth Systems, 2024
Modular Compositional Learning Improves 1D Hydrodynamic Lake Model Performance by Merging Process-Based Modeling With Deep LearningRobert Ladwig, Arka Daw, Ellen A Albright, and 6 more authorsJournal of Advances in Modeling Earth Systems, 2024Hybrid Knowledge-Guided Machine Learning (KGML) models, which are deep learning models that utilize scientific theory and process-based model simulations, have shown improved performance over their process-based counterparts for the simulation of water temperature and hydrodynamics. We highlight the modular compositional learning (MCL) methodology as a novel design choice for the development of hybrid KGML models in which the model is decomposed into modular sub-components that can be process-based models and/or deep learning models. We develop a hybrid MCL model that integrates a deep learning model into a modularized, process-based model. To achieve this, we first train individual deep learning models with the output of the process-based models. In a second step, we fine-tune one deep learning model with observed field data. In this study, we replaced process-based calculations of vertical diffusive transport with deep learning. Finally, this fine-tuned deep learning model is integrated into the process-based model, creating the hybrid MCL model with improved overall projections for water temperature dynamics compared to the original process-based model. We further compare the performance of the hybrid MCL model with the process-based model and two alternative deep learning models and highlight how the hybrid MCL model has the best performance for projecting water temperature, Schmidt stability, buoyancy frequency, and depths of different isotherms. Modular compositional learning can be applied to existing modularized, process-based model structures to make the projections more robust and improve model performance by letting deep learning estimate uncertain process calculations.
@article{ladwig2024modular, title = {Modular Compositional Learning Improves 1D Hydrodynamic Lake Model Performance by Merging Process-Based Modeling With Deep Learning}, author = {Ladwig, Robert and Daw, Arka and Albright, Ellen A and Buelo, Cal and Karpatne, Anuj and Meyer, Michael Frederick and Neog, Abhilash and Hanson, Paul C and Dugan, Hilary A}, journal = {Journal of Advances in Modeling Earth Systems}, volume = {16}, number = {1}, pages = {e2023MS003953}, year = {2024}, publisher = {Wiley Online Library}, category = {Conference Publications} }
2023
- ICML 2023
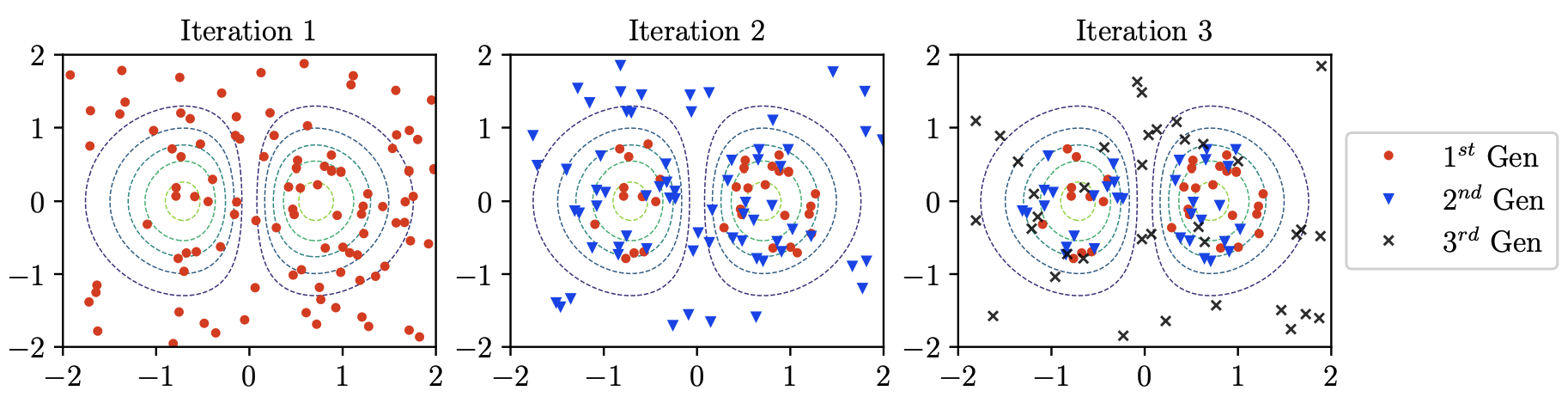 Mitigating propagation failures in physics-informed neural networks using retain-resample-release (r3) samplingArka Daw, Jie Bu, Sifan Wang, and 2 more authorsIn Proceedings of the 40th International Conference on Machine Learning, Honolulu, Hawaii, USA, 2023
Mitigating propagation failures in physics-informed neural networks using retain-resample-release (r3) samplingArka Daw, Jie Bu, Sifan Wang, and 2 more authorsIn Proceedings of the 40th International Conference on Machine Learning, Honolulu, Hawaii, USA, 2023Despite the success of physics-informed neural networks (PINNs) in approximating partial differential equations (PDEs), PINNs can sometimes fail to converge to the correct solution in problems involving complicated PDEs. This is reflected in several recent studies on characterizing the "failure modes" of PINNs, although a thorough understanding of the connection between PINN failure modes and sampling strategies is missing. In this paper, we provide a novel perspective of failure modes of PINNs by hypothesizing that training PINNs relies on successful "propagation" of solution from initial and/or boundary condition points to interior points. We show that PINNs with poor sampling strategies can get stuck at trivial solutions if there are propagation failures, characterized by highly imbalanced PDE residual fields. To mitigate propagation failures, we propose a novel Retain-Resample-Release sampling (R3) algorithm that can incrementally accumulate collocation points in regions of high PDE residuals with little to no computational overhead. We provide an extension of R3 sampling to respect the principle of causality while solving timedependent PDEs. We theoretically analyze the behavior of R3 sampling and empirically demonstrate its efficacy and efficiency in comparison with baselines on a variety of PDE problems.
@inproceedings{daw2022mitigating, author = {Daw, Arka and Bu, Jie and Wang, Sifan and Perdikaris, Paris and Karpatne, Anuj}, title = {Mitigating propagation failures in physics-informed neural networks using retain-resample-release (r3) sampling}, year = {2023}, publisher = {JMLR.org}, booktitle = {Proceedings of the 40th International Conference on Machine Learning}, articleno = {288}, numpages = {39}, location = {Honolulu, Hawaii, USA}, series = {ICML'23}, category = {Conference Publications} }
2022
- IEEE Big Data 2022
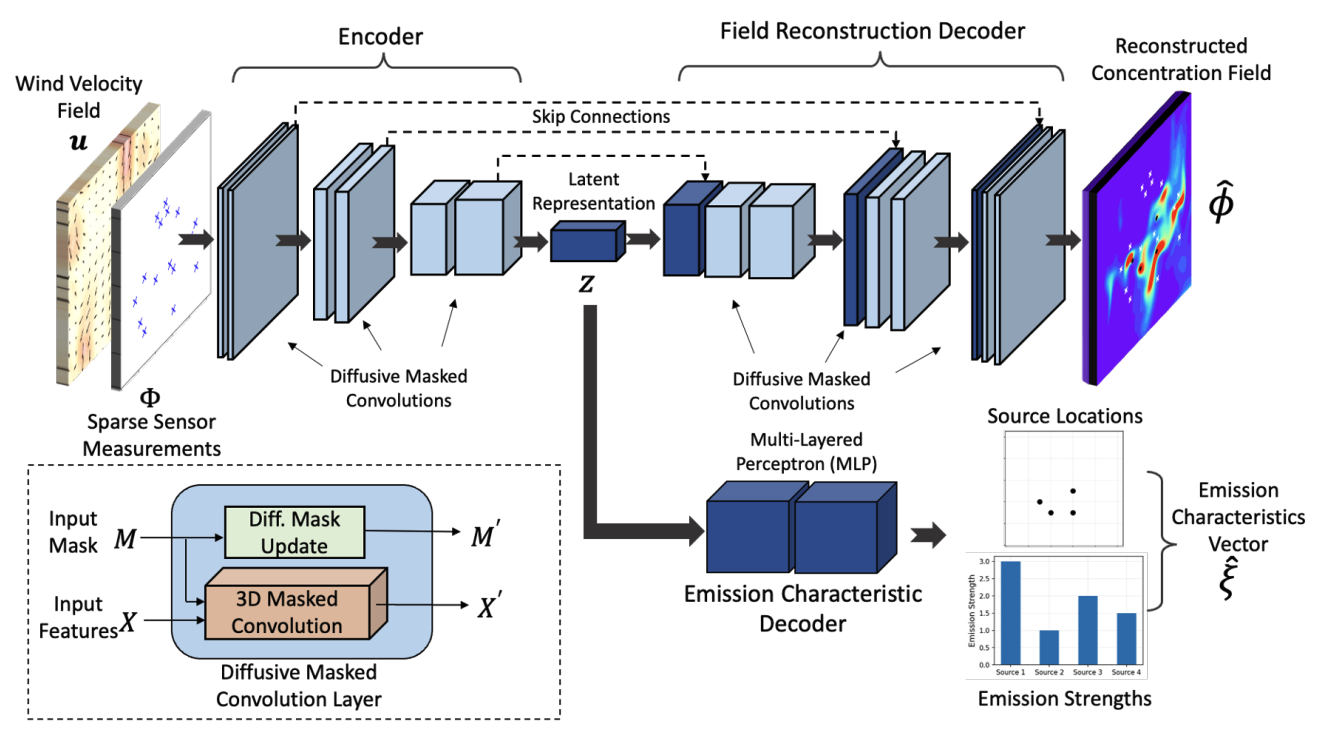 Multi-task learning for source attribution and field reconstruction for methane monitoringArka Daw, Kyongmin Yeo, Anuj Karpatne, and 1 more authorIn 2022 IEEE International Conference on Big Data (Big Data), 2022
Multi-task learning for source attribution and field reconstruction for methane monitoringArka Daw, Kyongmin Yeo, Anuj Karpatne, and 1 more authorIn 2022 IEEE International Conference on Big Data (Big Data), 2022Inferring the source information of greenhouse gases, such as methane, from spatially sparse sensor observations is an essential element in mitigating climate change. While it is well understood that the complex behavior of the atmospheric dispersion of such pollutants is governed by the Advection-Diffusion equation, it is difficult to directly apply the governing equations to identify the source location and magnitude (inverse problem) because of the spatially sparse and noisy observations, i.e., the pollution concentration is known only at the sensor locations and sensors sensitivity is limited. Here, we develop a multi-task learning framework that can provide high-fidelity reconstruction of the concentration field and identify emission characteristics of the pollution sources such as their location, emission strength, etc. from sparse sensor observations. We demonstrate that our proposed framework is able to achieve accurate reconstruction of the methane concentrations from sparse sensor measurements as well as precisely pin-point the location and emission strength of these pollution sources.
@inproceedings{daw2022multi, title = {Multi-task learning for source attribution and field reconstruction for methane monitoring}, author = {Daw, Arka and Yeo, Kyongmin and Karpatne, Anuj and Klein, Levente}, booktitle = {2022 IEEE International Conference on Big Data (Big Data)}, pages = {4835--4841}, year = {2022}, organization = {IEEE}, category = {Conference Publications} } - KGML Book
 Physics-guided neural networks (pgnn): An application in lake temperature modelingArka Daw, Anuj Karpatne, William D Watkins, and 2 more authorsIn Knowledge Guided Machine Learning, 2022
Physics-guided neural networks (pgnn): An application in lake temperature modelingArka Daw, Anuj Karpatne, William D Watkins, and 2 more authorsIn Knowledge Guided Machine Learning, 2022This chapter introduces a framework for combining scientific knowledge of physics-based models with neural networks to advance scientific discovery. It explains termed physics-guided neural networks (PGNN), leverages the output of physics-based model simulations along with observational features in a hybrid modeling setup to generate predictions using a neural network architecture. Data science has become an indispensable tool for knowledge discovery in the era of big data, as the volume of data continues to explode in practically every research domain. Recent advances in data science such as deep learning have been immensely successful in transforming the state-of-the-art in a number of commercial and industrial applications such as natural language translation and image classification, using billions or even trillions of data samples. Accurate water temperatures are critical to understanding contemporary change, and for predicting future thermal habitat of economically valuable fish.
@incollection{daw2022physicspgnn, title = {Physics-guided neural networks (pgnn): An application in lake temperature modeling}, author = {Daw, Arka and Karpatne, Anuj and Watkins, William D and Read, Jordan S and Kumar, Vipin}, booktitle = {Knowledge Guided Machine Learning}, pages = {353--372}, year = {2022}, publisher = {Chapman and Hall/CRC}, category = {Conference Publications} } - KGML Book
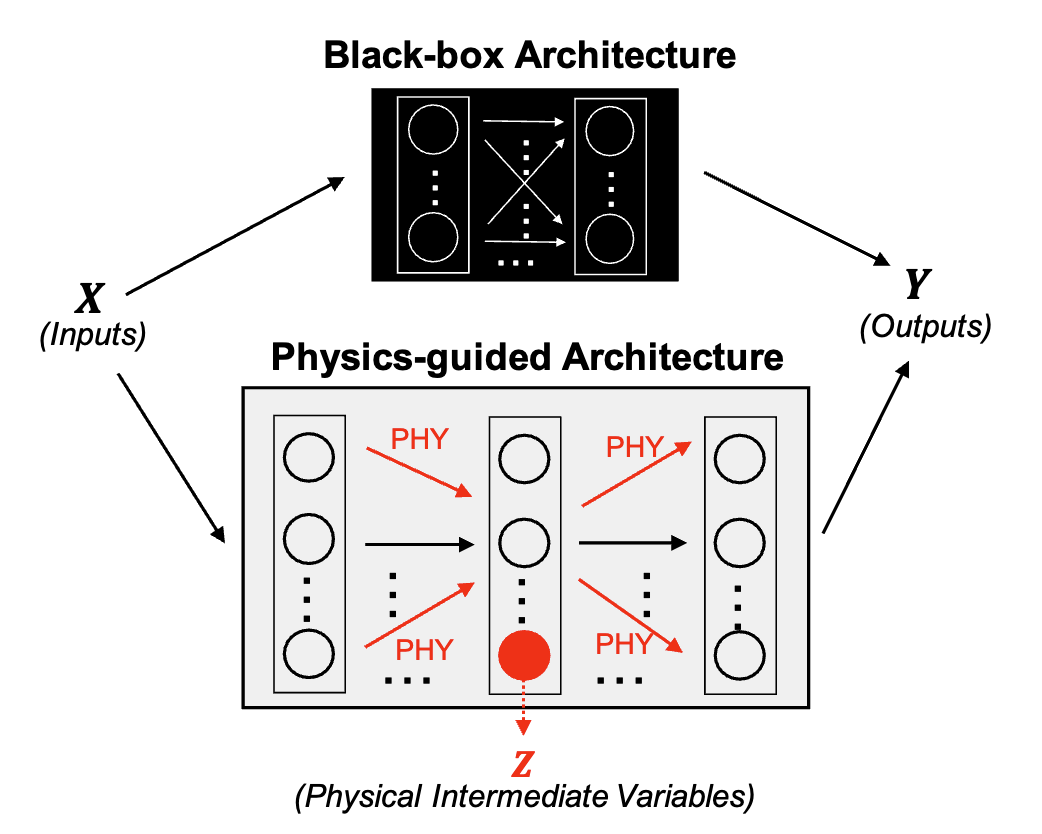 Physics-guided architecture (PGA) of LSTM models for uncertainty quantification in lake temperature modelingArka Daw, R Quinn Thomas, Cayelan C Carey, and 3 more authorsIn Knowledge Guided Machine Learning, 2022
Physics-guided architecture (PGA) of LSTM models for uncertainty quantification in lake temperature modelingArka Daw, R Quinn Thomas, Cayelan C Carey, and 3 more authorsIn Knowledge Guided Machine Learning, 2022This chapter focuses on meeting the need to produce neural network outputs that are physically consistent and also express uncertainties, a rare combination to date. It explains the effectiveness of physics-guided architecture - long-short-term-memory (PGA-LSTM) in achieving better generalizability and physical consistency over data collected from Lake Mendota in Wisconsin and Falling Creek Reservoir in Virginia, even with limited training data. Even though PGL formulations result in improvements in the generalization performance and lead to machine learning (ML) predictions that are more physically consistent, simply adding the physics-based loss function in the learning objective does not overcome the black-box nature of neural network architectures, which often involve arbitrary design choices. The temperature of water in a lake is a fundamental driver of lake biogeochemical processes, and it controls the growth, survival, and reproduction of fishes in the lake.
@incollection{daw2022physicspga, title = {Physics-guided architecture (PGA) of LSTM models for uncertainty quantification in lake temperature modeling}, author = {Daw, Arka and Thomas, R Quinn and Carey, Cayelan C and Read, Jordan S and Appling, Alison P and Karpatne, Anuj}, booktitle = {Knowledge Guided Machine Learning}, pages = {399--416}, year = {2022}, publisher = {Chapman and Hall/CRC}, category = {Conference Publications} }
2021
- NeurIPS 2021
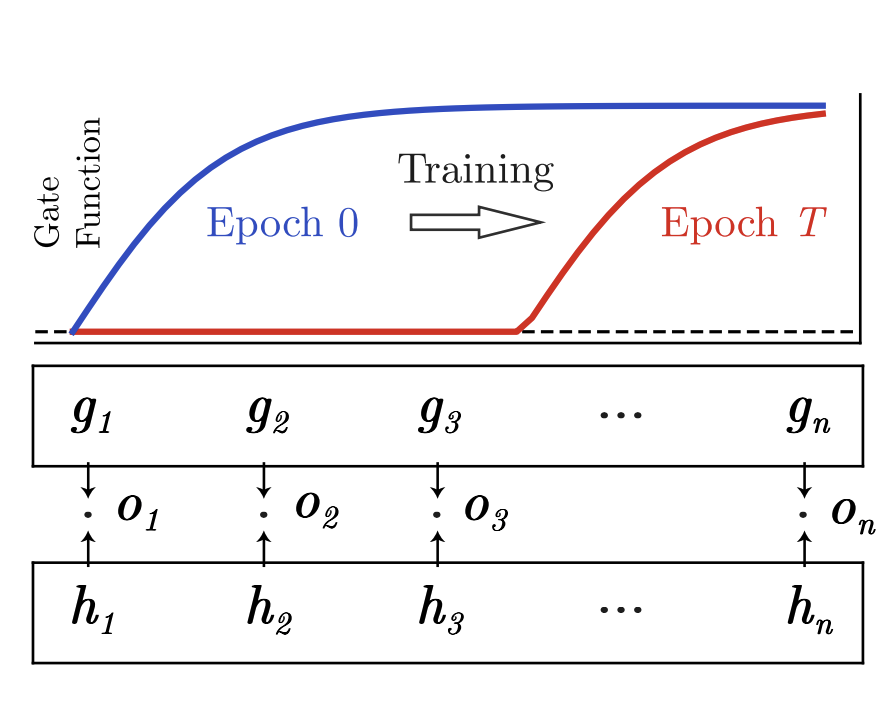 Learning compact representations of neural networks using discriminative masking (DAM)Jie Bu*, Arka Daw*, M Maruf*, and 1 more authorAdvances in Neural Information Processing Systems, 2021
Learning compact representations of neural networks using discriminative masking (DAM)Jie Bu*, Arka Daw*, M Maruf*, and 1 more authorAdvances in Neural Information Processing Systems, 2021A central goal in deep learning is to learn compact representations of features at every layer of a neural network, which is useful for both unsupervised representation learning and structured network pruning. While there is a growing body of work in structured pruning, current state-of-the-art methods suffer from two key limitations: (i) instability during training, and (ii) need for an additional step of fine-tuning, which is resource-intensive. At the core of these limitations is the lack of a systematic approach that jointly prunes and refines weights during training in a single stage, and does not require any fine-tuning upon convergence to achieve state-of-the-art performance. We present a novel single-stage structured pruning method termed DiscriminAtive Masking (DAM). The key intuition behind DAM is to discriminatively prefer some of the neurons to be refined during the training process, while gradually masking out other neurons. We show that our proposed DAM approach has remarkably good performance over a diverse range of applications in representation learning and structured pruning, including dimensionality reduction, recommendation system, graph representation learning, and structured pruning for image classification. We also theoretically show that the learning objective of DAM is directly related to minimizing the L_0 norm of the masking layer. All of our codes and datasets are available https://github.com/jayroxis/dam-pytorch.
@article{bu2021learning, title = {Learning compact representations of neural networks using discriminative masking (DAM)}, author = {Bu, Jie and Daw, Arka and Maruf, M and Karpatne, Anuj}, journal = {Advances in Neural Information Processing Systems}, volume = {34}, pages = {3491--3503}, year = {2021}, publisher = {NeurIPS}, category = {Conference Publications} } - KDD 2021
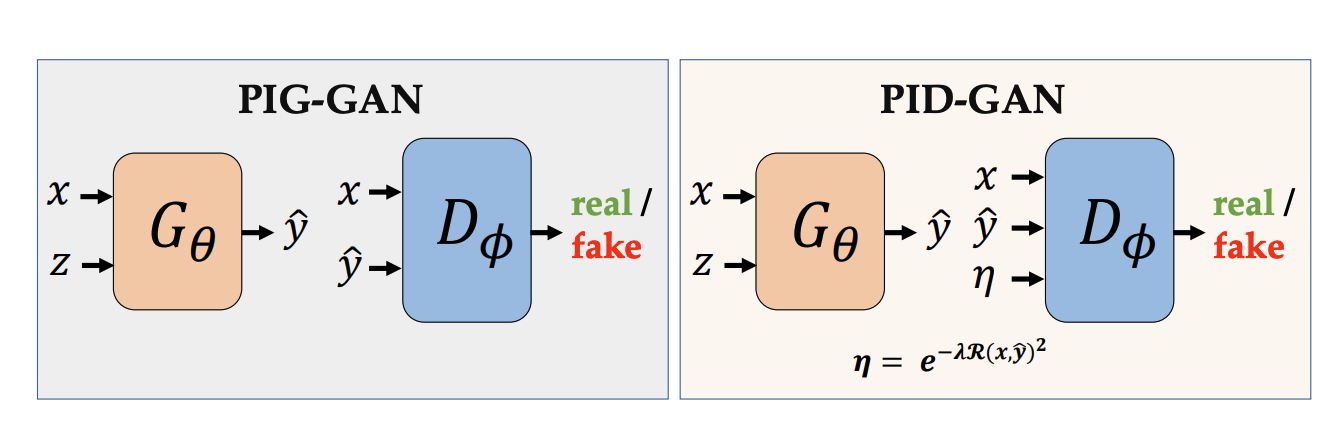 PID-GAN: A GAN Framework based on a Physics-informed Discriminator for Uncertainty Quantification with PhysicsArka Daw, M. Maruf, and Anuj KarpatneIn Proceedings of the 27th ACM SIGKDD Conference on Knowledge Discovery & Data Mining, Virtual Event, Singapore, 2021
PID-GAN: A GAN Framework based on a Physics-informed Discriminator for Uncertainty Quantification with PhysicsArka Daw, M. Maruf, and Anuj KarpatneIn Proceedings of the 27th ACM SIGKDD Conference on Knowledge Discovery & Data Mining, Virtual Event, Singapore, 2021As applications of deep learning (DL) continue to seep into critical scientific use-cases, the importance of performing uncertainty quantification (UQ) with DL has become more pressing than ever before. In scientific applications, it is also important to inform the learning of DL models with knowledge of physics of the problem to produce physically consistent and generalized solutions. This is referred to as the emerging field of physics-informed deep learning (PIDL). We consider the problem of developing PIDL formulations that can also perform UQ. To this end, we propose a novel physics-informed GAN architecture, termed PID-GAN, where the knowledge of physics is used to inform the learning of both the generator and discriminator models, making ample use of unlabeled data instances. We show that our proposed PID-GAN framework does not suffer from imbalance of generator gradients from multiple loss terms as compared to state-of-the-art. We also empirically demonstrate the efficacy of our proposed framework on a variety of case studies involving benchmark physics-based PDEs as well as imperfect physics. All the code and datasets used in this study have been made available on this link: https://github.com/arkadaw9/PID-GAN.
@inproceedings{daw2021pidgan, author = {Daw, Arka and Maruf, M. and Karpatne, Anuj}, title = {PID-GAN: A GAN Framework based on a Physics-informed Discriminator for Uncertainty Quantification with Physics}, year = {2021}, isbn = {9781450383325}, publisher = {Association for Computing Machinery}, address = {New York, NY, USA}, url = {https://doi.org/10.1145/3447548.3467449}, doi = {10.1145/3447548.3467449}, booktitle = {Proceedings of the 27th ACM SIGKDD Conference on Knowledge Discovery \& Data Mining}, pages = {237–247}, numpages = {11}, keywords = {uncertainty quantification, physics-informed neural networks, generative adversarial networks}, location = {Virtual Event, Singapore}, series = {KDD '21}, category = {Conference Publications} }
2020
- SDM 2020
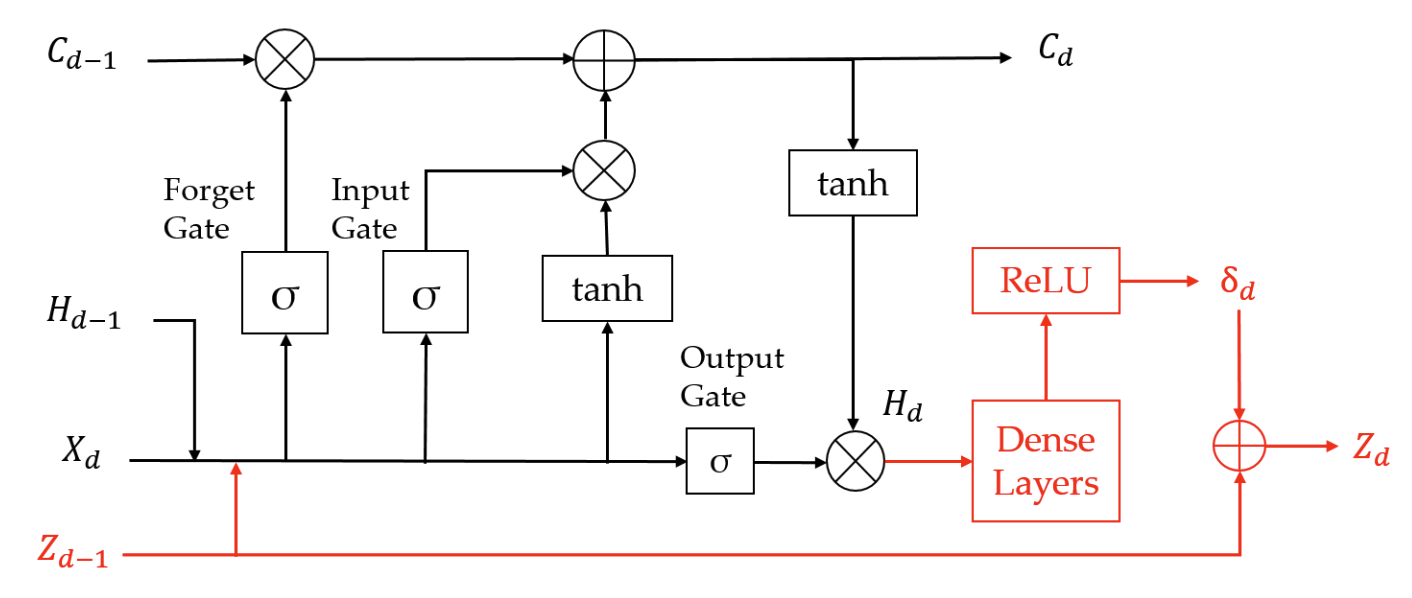 Physics-guided architecture (pga) of neural networks for quantifying uncertainty in lake temperature modelingArka Daw, R Quinn Thomas, Cayelan C Carey, and 3 more authorsIn Proceedings of the 2020 siam international conference on data mining, 2020
Physics-guided architecture (pga) of neural networks for quantifying uncertainty in lake temperature modelingArka Daw, R Quinn Thomas, Cayelan C Carey, and 3 more authorsIn Proceedings of the 2020 siam international conference on data mining, 2020To simultaneously address the rising need of expressing uncertainties in deep learning models along with producing model outputs which are consistent with the known scientific knowledge, we propose a novel physics-guided architecture (PGA) of neural networks in the context of lake temperature modeling where the physical constraints are hard coded in the neural network architecture. This allows us to integrate such models with state of the art uncertainty estimation approaches such as Monte Carlo (MC) Dropout without sacrificing the physical consistency of our results. We demonstrate the effectiveness of our approach in ensuring better generalizability as well as physical consistency in MC estimates over data collected from Lake Mendota in Wisconsin and Falling Creek Reservoir in Virginia, even with limited training data. We further show that our MC estimates correctly match the distribution of ground-truth observations, thus making the PGA paradigm amenable to physically grounded uncertainty quantification.
@inproceedings{daw2020physict, title = {Physics-guided architecture (pga) of neural networks for quantifying uncertainty in lake temperature modeling}, author = {Daw, Arka and Thomas, R Quinn and Carey, Cayelan C and Read, Jordan S and Appling, Alison P and Karpatne, Anuj}, booktitle = {Proceedings of the 2020 siam international conference on data mining}, pages = {532--540}, year = {2020}, organization = {SIAM}, category = {Conference Publications} }
Preprints
2024
- Arxiv
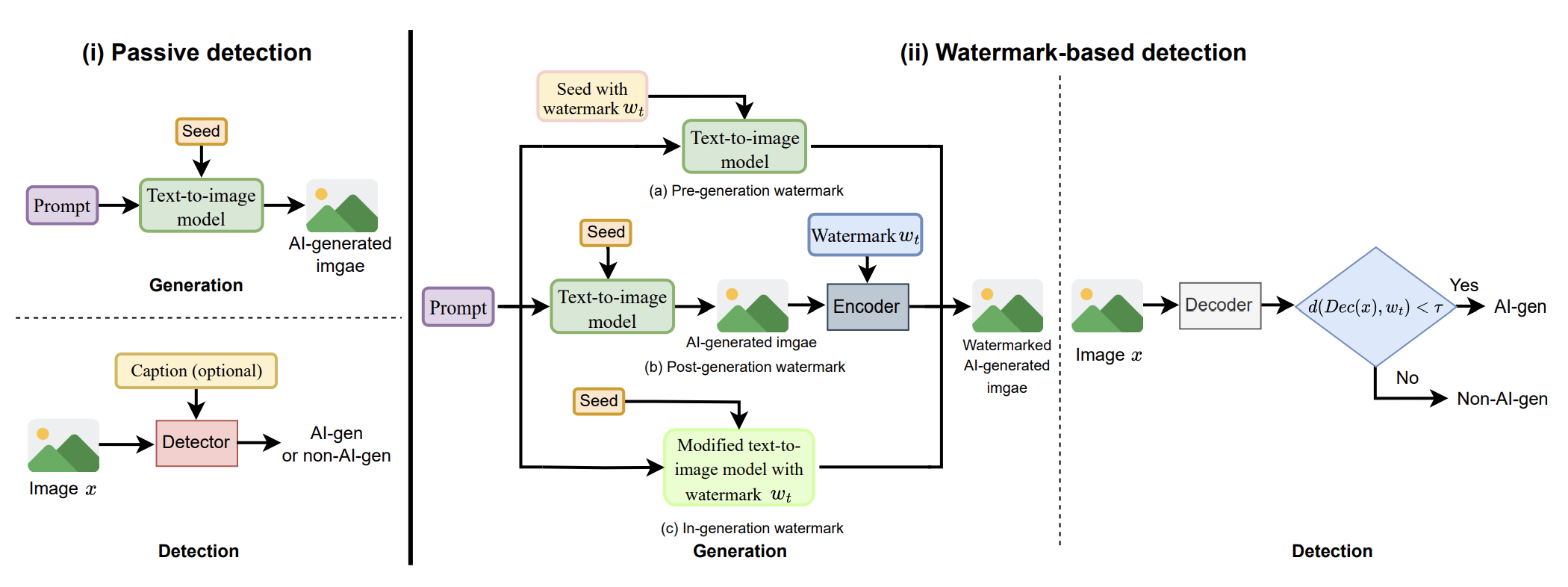 AI-generated Image Detection: Passive or Watermark?Moyang Guo, Yuepeng Hu, Zhengyuan Jiang, and 4 more authorsarXiv preprint arXiv:2411.13553, 2024
AI-generated Image Detection: Passive or Watermark?Moyang Guo, Yuepeng Hu, Zhengyuan Jiang, and 4 more authorsarXiv preprint arXiv:2411.13553, 2024While text-to-image models offer numerous benefits, they also pose significant societal risks. Detecting AI-generated images is crucial for mitigating these risks. Detection methods can be broadly categorized into passive and watermark-based approaches: passive detectors rely on artifacts present in AI-generated images, whereas watermark-based detectors proactively embed watermarks into such images. A key question is which type of detector performs better in terms of effectiveness, robustness, and efficiency. However, the current literature lacks a comprehensive understanding of this issue. In this work, we aim to bridge that gap by developing ImageDetectBench, the first comprehensive benchmark to compare the effectiveness, robustness, and efficiency of passive and watermark-based detectors. Our benchmark includes four datasets, each containing a mix of AI-generated and non-AI-generated images. We evaluate five passive detectors and four watermark-based detectors against eight types of common perturbations and three types of adversarial perturbations. Our benchmark results reveal several interesting findings. For instance, watermark-based detectors consistently outperform passive detectors, both in the presence and absence of perturbations. Based on these insights, we provide recommendations for detecting AI-generated images, e.g., when both types of detectors are applicable, watermark-based detectors should be the preferred choice.
@article{guo2024ai, title = {AI-generated Image Detection: Passive or Watermark?}, author = {Guo, Moyang and Hu, Yuepeng and Jiang, Zhengyuan and Li, Zeyu and Sadovnik, Amir and Daw, Arka and Gong, Neil}, journal = {arXiv preprint arXiv:2411.13553}, year = {2024}, publisher = {Arxiv}, category = {Preprints} } - Arxiv
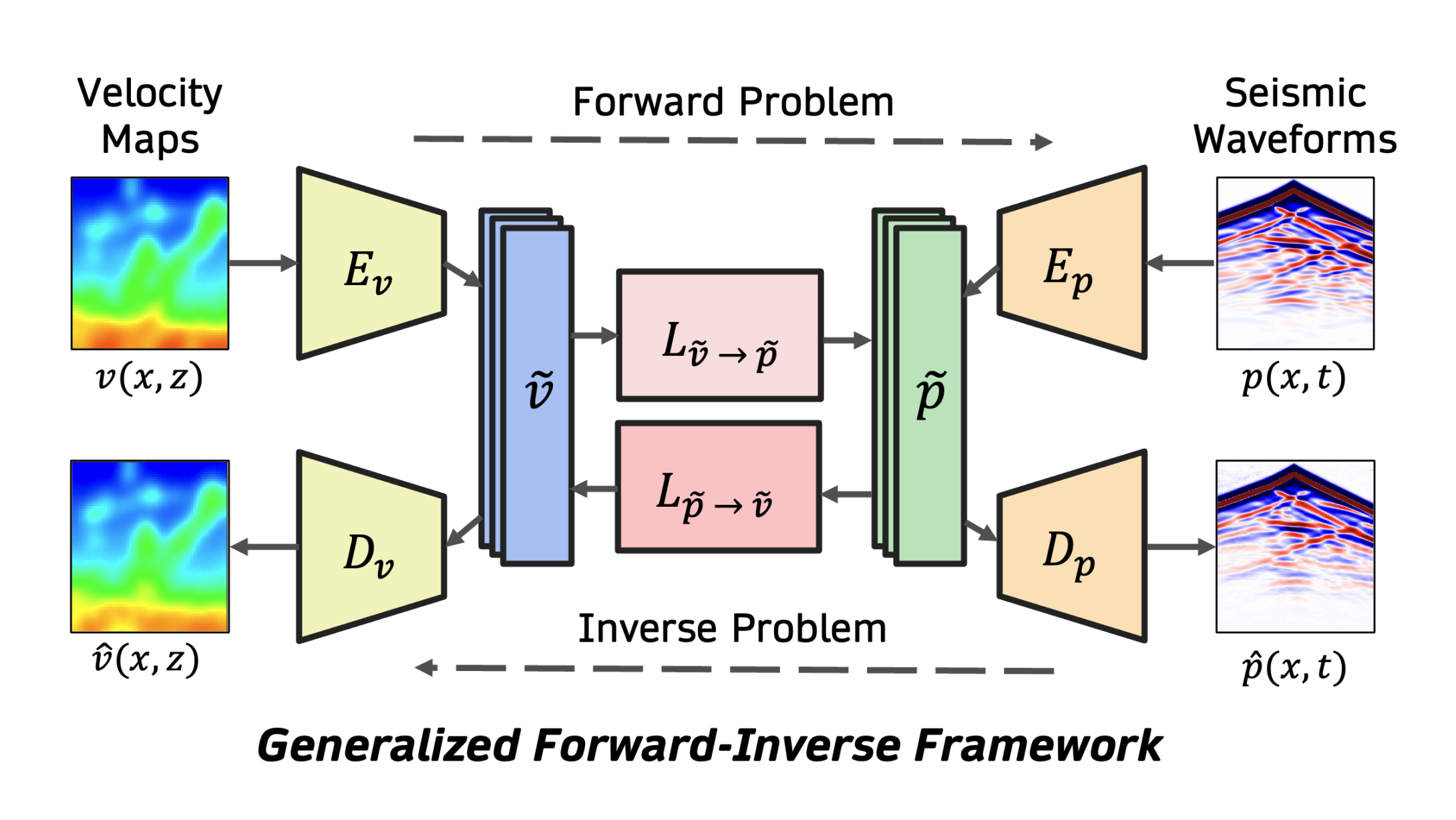 A Unified Framework for Forward and Inverse Problems in Subsurface Imaging using Latent Space TranslationsNaveen Gupta*, Medha Sawhney*, Arka Daw*, and 2 more authorsarXiv preprint arXiv:2410.11247, 2024
A Unified Framework for Forward and Inverse Problems in Subsurface Imaging using Latent Space TranslationsNaveen Gupta*, Medha Sawhney*, Arka Daw*, and 2 more authorsarXiv preprint arXiv:2410.11247, 2024In subsurface imaging, learning the mapping from velocity maps to seismic waveforms (forward problem) and waveforms to velocity (inverse problem) is important for several applications. While traditional techniques for solving forward and inverse problems are computationally prohibitive, there is a growing interest in leveraging recent advances in deep learning to learn the mapping between velocity maps and seismic waveform images directly from data. Despite the variety of architectures explored in previous works, several open questions still remain unanswered such as the effect of latent space sizes, the importance of manifold learning, the complexity of translation models, and the value of jointly solving forward and inverse problems. We propose a unified framework to systematically characterize prior research in this area termed the Generalized Forward-Inverse (GFI) framework, building on the assumption of manifolds and latent space translations. We show that GFI encompasses previous works in deep learning for subsurface imaging, which can be viewed as specific instantiations of GFI. We also propose two new model architectures within the framework of GFI: Latent U-Net and Invertible X-Net, leveraging the power of U-Nets for domain translation and the ability of IU-Nets to simultaneously learn forward and inverse translations, respectively. We show that our proposed models achieve state-of-the-art (SOTA) performance for forward and inverse problems on a wide range of synthetic datasets, and also investigate their zero-shot effectiveness on two real-world-like datasets.
@article{gupta2024unified, title = {A Unified Framework for Forward and Inverse Problems in Subsurface Imaging using Latent Space Translations}, author = {Gupta, Naveen and Sawhney, Medha and Daw, Arka and Lin, Youzuo and Karpatne, Anuj}, journal = {arXiv preprint arXiv:2410.11247}, year = {2024}, publisher = {Arxiv}, category = {Preprints} } - Arxiv
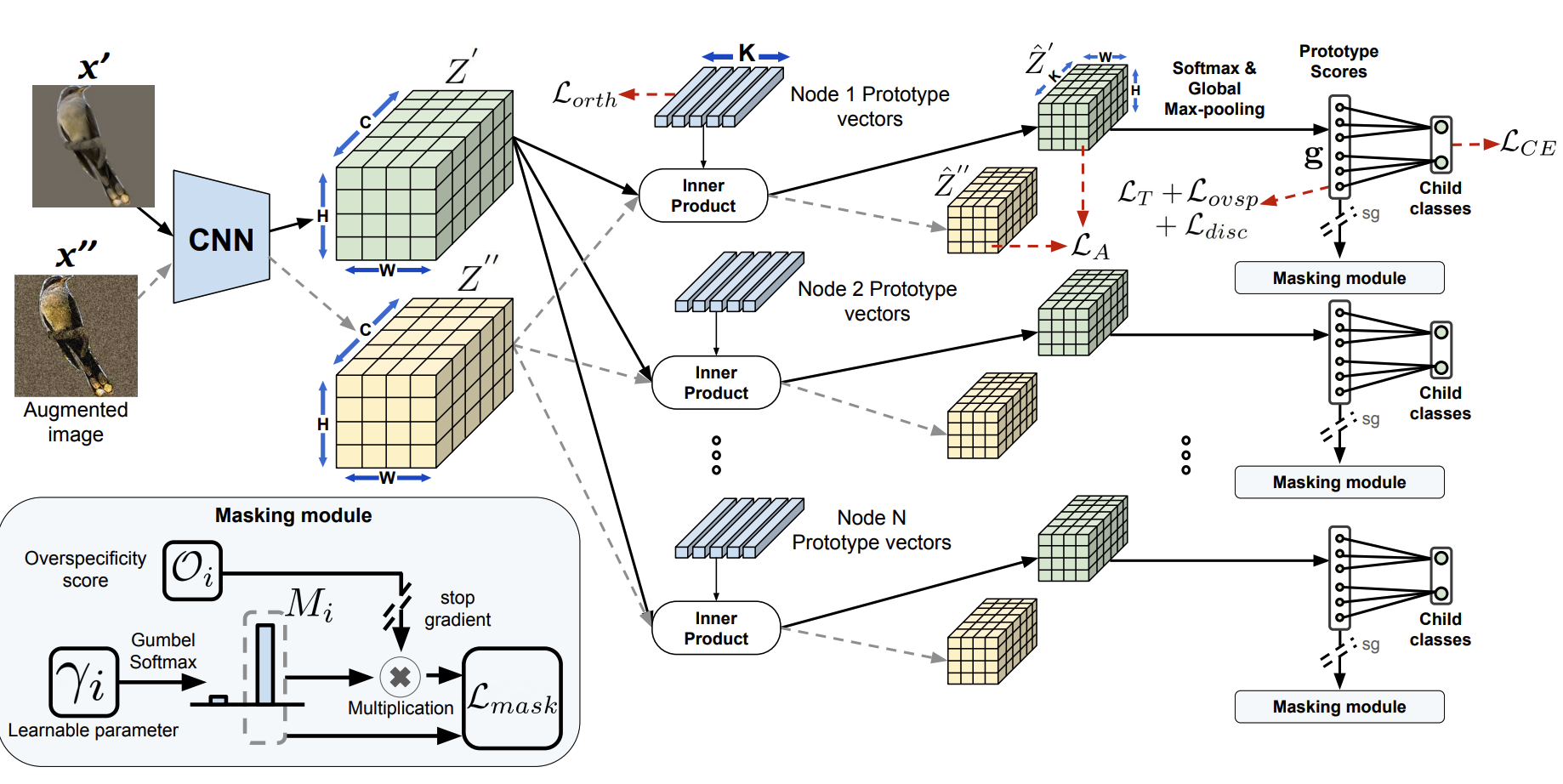 What Do You See in Common? Learning Hierarchical Prototypes over Tree-of-Life to Discover Evolutionary TraitsHarish Babu Manogaran, M Maruf, Arka Daw, and 8 more authorsarXiv preprint arXiv:2409.02335, 2024
What Do You See in Common? Learning Hierarchical Prototypes over Tree-of-Life to Discover Evolutionary TraitsHarish Babu Manogaran, M Maruf, Arka Daw, and 8 more authorsarXiv preprint arXiv:2409.02335, 2024A grand challenge in biology is to discover evolutionary traits - features of organisms common to a group of species with a shared ancestor in the tree of life (also referred to as phylogenetic tree). With the growing availability of image repositories in biology, there is a tremendous opportunity to discover evolutionary traits directly from images in the form of a hierarchy of prototypes. However, current prototype-based methods are mostly designed to operate over a flat structure of classes and face several challenges in discovering hierarchical prototypes, including the issue of learning over-specific features at internal nodes. To overcome these challenges, we introduce the framework of Hierarchy aligned Commonality through Prototypical Networks (HComP-Net). We empirically show that HComP-Net learns prototypes that are accurate, semantically consistent, and generalizable to unseen species in comparison to baselines on birds, butterflies, and fishes datasets.
@article{manogaran2024you, title = {What Do You See in Common? Learning Hierarchical Prototypes over Tree-of-Life to Discover Evolutionary Traits}, author = {Manogaran, Harish Babu and Maruf, M and Daw, Arka and Mehrab, Kazi Sajeed and Charpentier, Caleb Patrick and Uyeda, Josef C and Dahdul, Wasila and Thompson, Matthew J and Campolongo, Elizabeth G and Provost, Kaiya L and others}, journal = {arXiv preprint arXiv:2409.02335}, year = {2024}, publisher = {Arxiv}, category = {Preprints} } - Arxiv
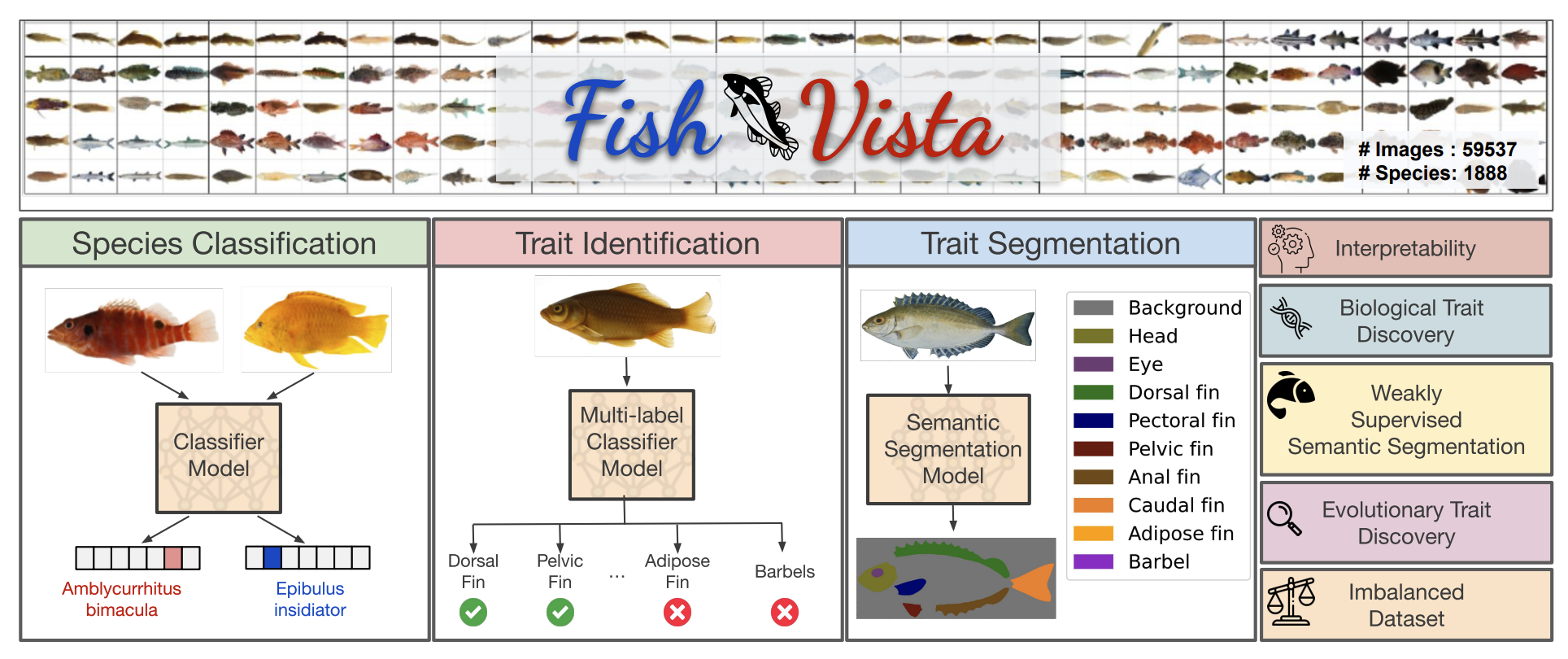 Fish-Vista: A Multi-Purpose Dataset for Understanding & Identification of Traits from ImagesKazi Sajeed Mehrab*, M Maruf*, Arka Daw*, and 8 more authorsarXiv preprint arXiv:2407.08027, 2024
Fish-Vista: A Multi-Purpose Dataset for Understanding & Identification of Traits from ImagesKazi Sajeed Mehrab*, M Maruf*, Arka Daw*, and 8 more authorsarXiv preprint arXiv:2407.08027, 2024Fishes are integral to both ecological systems and economic sectors, and studying fish traits is crucial for understanding biodiversity patterns and macro-evolution trends. To enable the analysis of visual traits from fish images, we introduce the Fish-Visual Trait Analysis (Fish-Vista) dataset - a large, annotated collection of about 60K fish images spanning 1900 different species, supporting several challenging and biologically relevant tasks including species classification, trait identification, and trait segmentation. These images have been curated through a sophisticated data processing pipeline applied to a cumulative set of images obtained from various museum collections. Fish-Vista provides fine-grained labels of various visual traits present in each image. It also offers pixel-level annotations of 9 different traits for 2427 fish images, facilitating additional trait segmentation and localization tasks. The ultimate goal of Fish-Vista is to provide a clean, carefully curated, high-resolution dataset that can serve as a foundation for accelerating biological discoveries using advances in AI. Finally, we provide a comprehensive analysis of state-of-the-art deep learning techniques on Fish-Vista.
@article{mehrab2024fish, title = {Fish-Vista: A Multi-Purpose Dataset for Understanding \& Identification of Traits from Images}, author = {Mehrab, Kazi Sajeed and Maruf, M and Daw, Arka and Manogaran, Harish Babu and Neog, Abhilash and Khurana, Mridul and Altintas, Bahadir and Bakis, Yasin and Campolongo, Elizabeth G and Thompson, Matthew J and others}, journal = {arXiv preprint arXiv:2407.08027}, year = {2024}, publisher = {Arxiv}, category = {Preprints} }
2023
- Arxiv
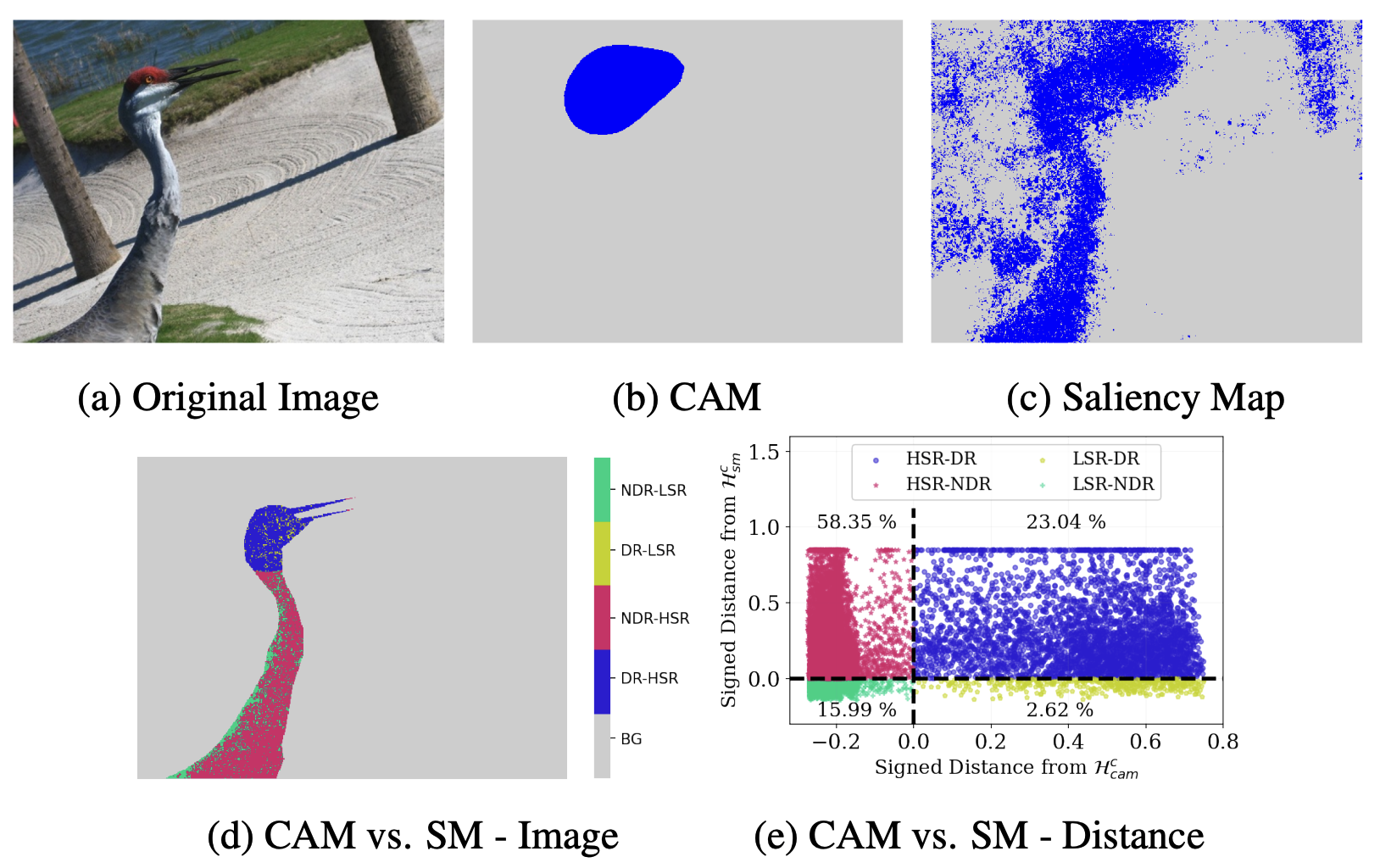 Beyond Discriminative Regions: Saliency Maps as Alternatives to CAMs for Weakly Supervised Semantic SegmentationM Maruf, Arka Daw, Amartya Dutta, and 2 more authorsarXiv preprint arXiv:2308.11052, 2023
Beyond Discriminative Regions: Saliency Maps as Alternatives to CAMs for Weakly Supervised Semantic SegmentationM Maruf, Arka Daw, Amartya Dutta, and 2 more authorsarXiv preprint arXiv:2308.11052, 2023In recent years, several Weakly Supervised Semantic Segmentation (WS3) methods have been proposed that use class activation maps (CAMs) generated by a classifier to produce pseudo-ground truths for training segmentation models. While CAMs are good at highlighting discriminative regions (DR) of an image, they are known to disregard regions of the object that do not contribute to the classifier’s prediction, termed non-discriminative regions (NDR). In contrast, attribution methods such as saliency maps provide an alternative approach for assigning a score to every pixel based on its contribution to the classification prediction. This paper provides a comprehensive comparison between saliencies and CAMs for WS3. Our study includes multiple perspectives on understanding their similarities and dissimilarities. Moreover, we provide new evaluation metrics that perform a comprehensive assessment of WS3 performance of alternative methods w.r.t. CAMs. We demonstrate the effectiveness of saliencies in addressing the limitation of CAMs through our empirical studies on benchmark datasets. Furthermore, we propose random cropping as a stochastic aggregation technique that improves the performance of saliency, making it a strong alternative to CAM for WS3.
@article{maruf2023beyond, title = {Beyond Discriminative Regions: Saliency Maps as Alternatives to CAMs for Weakly Supervised Semantic Segmentation}, author = {Maruf, M and Daw, Arka and Dutta, Amartya and Bu, Jie and Karpatne, Anuj}, journal = {arXiv preprint arXiv:2308.11052}, year = {2023}, publisher = {Arxiv}, category = {Preprints} }
2022
- BioRxiv
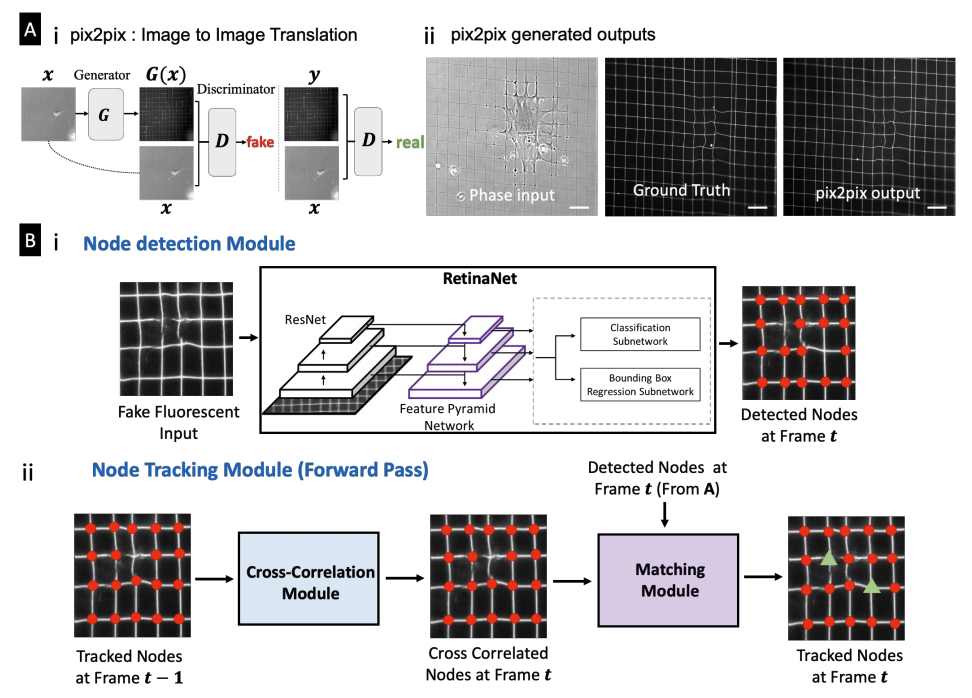 Deep Learning Enabled Label-free Cell Force Computation in Deformable Fibrous EnvironmentsAbinash Padhi*, Arka Daw*, Medha Sawhney, and 5 more authorsbioRxiv, 2022
Deep Learning Enabled Label-free Cell Force Computation in Deformable Fibrous EnvironmentsAbinash Padhi*, Arka Daw*, Medha Sawhney, and 5 more authorsbioRxiv, 2022Through force exertion, cells actively engage with their immediate fibrous extracellular matrix (ECM) environment, causing dynamic remodeling of the environment and influencing cellular shape and contractility changes in a feedforward loop. Controlling cell shapes and quantifying the force-driven dynamic reciprocal interactions in a label-free setting is vital to understand cell behavior in fibrous environments but currently unavailable. Here, we introduce a force measurement platform termed crosshatch nanonet force microscopy (cNFM) that reveals new insights into cell shape-force coupling. Using a suspended crosshatch network of fibers capable of recovering in vivo cell shapes, we utilize deep learning methods to circumvent the fiduciary fluorescent markers required to recognize fiber intersections. Our method provides high fidelity computer reconstruction of different fiber architectures by automatically translating phase-contrast time-lapse images into synthetic fluorescent images. An inverse problem based on the nonlinear mechanics of fiber networks is formulated to match the network deformation and deformed fiber shapes to estimate the forces. We reveal an order-of-magnitude force changes associated with cell shape changes during migration, forces during cell-cell interactions and force changes as single mesenchymal stem cells undergo differentiation. Overall, deep learning methods are employed in detecting and tracking highly compliant backgrounds to develop an automatic and label-free force measurement platform to describe cell shape-force coupling in fibrous environments that cells would likely interact with in vivo.
@article{padhi2022deep, title = {Deep Learning Enabled Label-free Cell Force Computation in Deformable Fibrous Environments}, author = {Padhi, Abinash and Daw, Arka and Sawhney, Medha and Talukder, Maahi M and Agashe, Atharva and Kale, Sohan and Karpatne, Anuj and Nain, Amrinder S}, journal = {bioRxiv}, pages = {2022--10}, year = {2022}, publisher = {Cold Spring Harbor Laboratory}, category = {Preprints} }
Workshop Papers
2024
- NeurIPS 2024
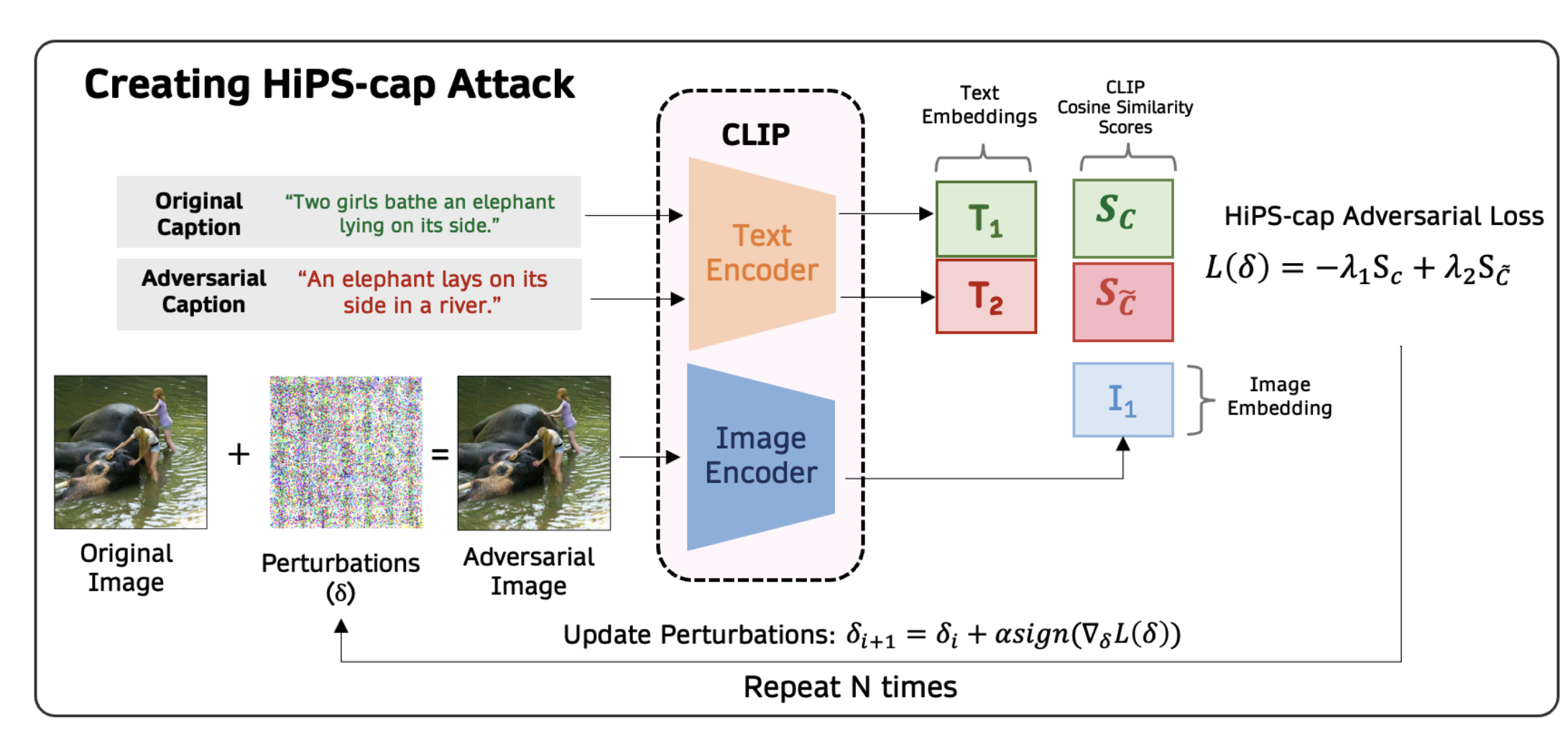 Hiding-in-Plain-Sight (HiPS) Attack on CLIP for Targetted Object Removal from ImagesArka Daw, Megan Hong-Thanh Chung, Maria Mahbub, and 1 more authorNeurIPS Workshop on New Frontiers in Adversarial Machine Learning, 2024
Hiding-in-Plain-Sight (HiPS) Attack on CLIP for Targetted Object Removal from ImagesArka Daw, Megan Hong-Thanh Chung, Maria Mahbub, and 1 more authorNeurIPS Workshop on New Frontiers in Adversarial Machine Learning, 2024Machine learning models are known to be vulnerable to adversarial attacks, but traditional attacks have mostly focused on single-modalities. With the rise of large multi-modal models (LMMs) like CLIP, which combine vision and language capabilities, new vulnerabilities have emerged. However, prior work in multimodal targeted attacks aim to completely change the model’s output to what the adversary wants. In many realistic scenarios, an adversary might seek to make only subtle modifications to the output, so that the changes go unnoticed by downstream models or even by humans. We introduce Hiding-in-Plain-Sight (HiPS) attacks, a novel class of adversarial attacks that subtly modifies model predictions by selectively concealing target object(s), as if the target object was absent from the scene. We propose two HiPS attack variants, HiPS-cls and HiPS-cap, and demonstrate their effectiveness in transferring to downstream image captioning models, such as CLIP-Cap, for targeted object removal from image captions.
@article{daw2024hiding, title = {Hiding-in-Plain-Sight (HiPS) Attack on CLIP for Targetted Object Removal from Images}, author = {Daw, Arka and Chung, Megan Hong-Thanh and Mahbub, Maria and Sadovnik, Amir}, journal = {NeurIPS Workshop on New Frontiers in Adversarial Machine Learning}, year = {2024}, publisher = {NeurIPS}, category = {Workshop Papers} } - NeurIPS 2024
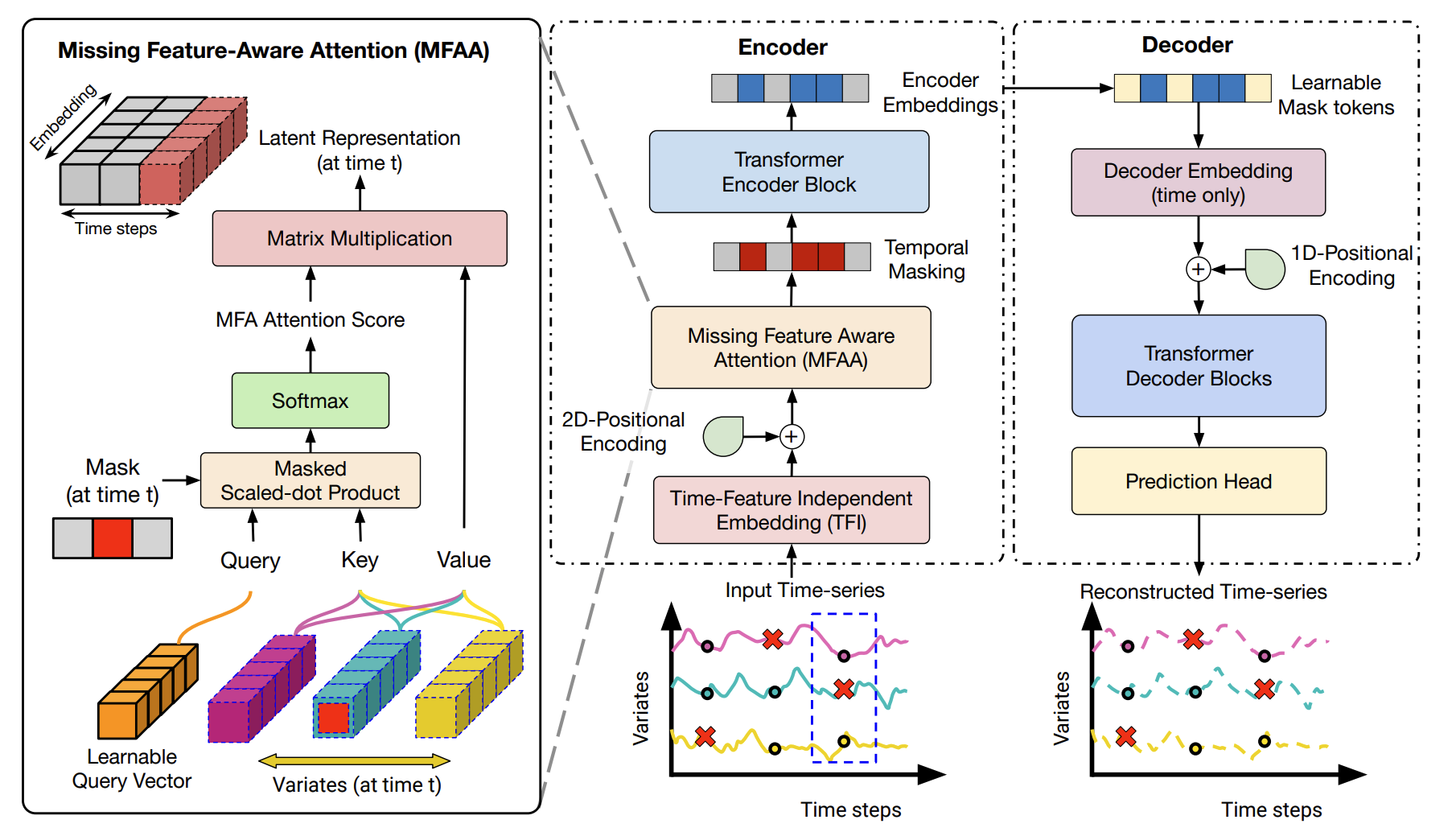 Masking the Gaps: An Imputation-Free Approach to Time Series Modeling with Missing DataAbhilash Neog, Arka Daw, Sepideh Fatemi Khorasgani, and 1 more authorIn NeurIPS Workshop on Time Series in the Age of Large Models, 2024
Masking the Gaps: An Imputation-Free Approach to Time Series Modeling with Missing DataAbhilash Neog, Arka Daw, Sepideh Fatemi Khorasgani, and 1 more authorIn NeurIPS Workshop on Time Series in the Age of Large Models, 2024A significant challenge in time-series (TS) modelling is presence of missing values in real-world TS datasets. Traditional two-stage frameworks, involving imputation followed by modeling, suffer from two key drawbacks: (1) the propagation of imputation errors into subsequent TS modeling, (2) the trade-offs between imputation efficacy and imputation complexity. While one-stage approaches attempt to address these limitations, they often struggle with scalability or fully leveraging partially observed features. To this end, we propose a novel imputation-free approach for handling missing values in time series termed \textbfMissing Feature-aware \textbfTime \textbfSeries \textbfModeling (\textbfMissTSM) with two main innovations. \textitFirst, we develop a novel embedding scheme that treats every combination of time-step and feature (or channel) as a distinct token. \textitSecond, we introduce a novel \textitMissing Feature-Aware Attention (MFAA) Layer to learn latent representations at every time-step based on partially observed features. We evaluate the effectiveness of MissTSM in handling missing values over multiple benchmark datasets.
@inproceedings{neogmasking, title = {Masking the Gaps: An Imputation-Free Approach to Time Series Modeling with Missing Data}, author = {Neog, Abhilash and Daw, Arka and Khorasgani, Sepideh Fatemi and Karpatne, Anuj}, booktitle = {NeurIPS Workshop on Time Series in the Age of Large Models}, year = {2024}, publisher = {NeurIPS}, category = {Workshop Papers} } - ICML 2024
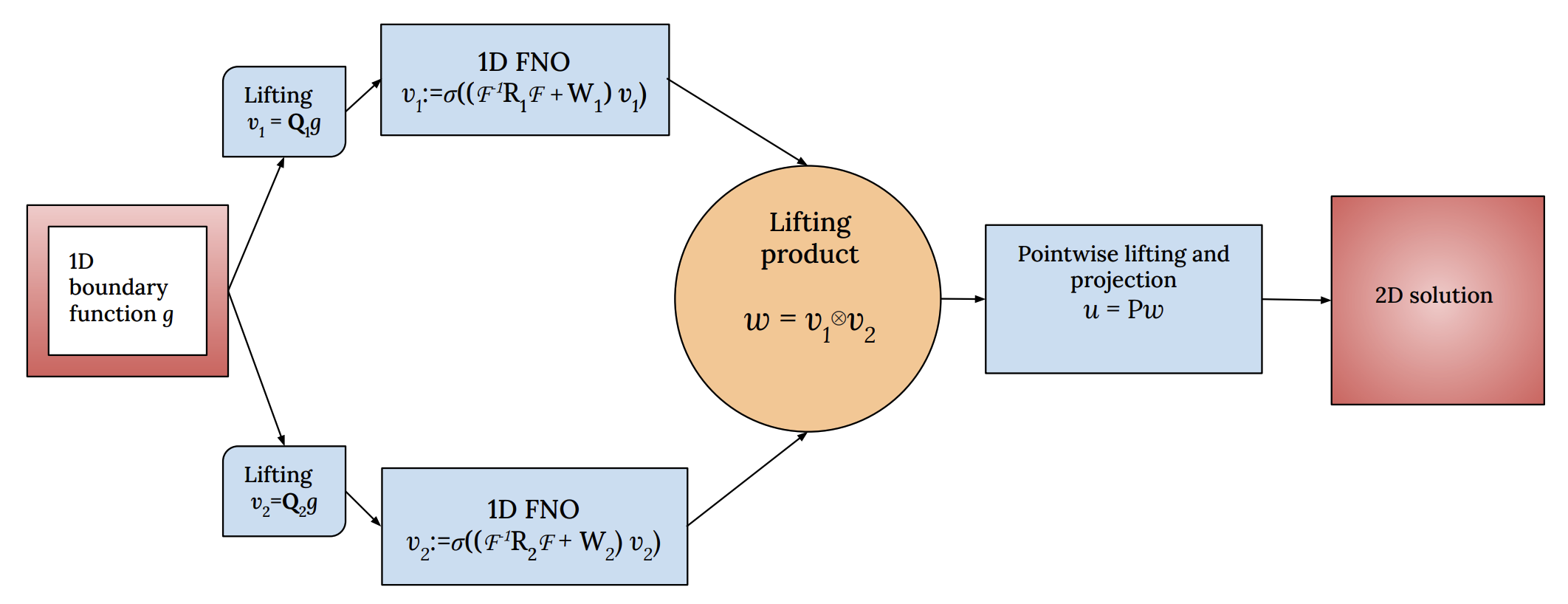 Learning the boundary-to-domain mapping using Lifting Product Fourier Neural Operators for partial differential equationsAditya Kashi, Arka Daw, Muralikrishnan Gopalakrishnan Meena, and 1 more authorAI4Science Workshop at International Conference on Machine Learning (ICML) 2024, 2024
Learning the boundary-to-domain mapping using Lifting Product Fourier Neural Operators for partial differential equationsAditya Kashi, Arka Daw, Muralikrishnan Gopalakrishnan Meena, and 1 more authorAI4Science Workshop at International Conference on Machine Learning (ICML) 2024, 2024Neural operators such as the Fourier Neural Operator (FNO) have been shown to provide resolution-independent deep learning models that can learn mappings between function spaces. For example, an initial condition can be mapped to the solution of a partial differential equation (PDE) at a future time-step using a neural operator. Despite the popularity of neural operators, their use to predict solution functions over a domain given only data over the boundary (such as a spatially varying Dirichlet boundary condition) remains unexplored. In this paper, we refer to such problems as boundary-to-domain problems; they have a wide range of applications in areas such as fluid mechanics, solid mechanics, heat transfer etc. We present a novel FNO-based architecture, named Lifting Product FNO (or LP-FNO) which can map arbitrary boundary functions defined on the lower-dimensional boundary to a solution in the entire domain. Specifically, two FNOs defined on the lower-dimensional boundary are lifted into the higher dimensional domain using our proposed lifting product layer. We demonstrate the efficacy and resolution independence of the proposed LP-FNO for the 2D Poisson equation.
@article{kashi2024learning, title = {Learning the boundary-to-domain mapping using Lifting Product Fourier Neural Operators for partial differential equations}, author = {Kashi, Aditya and Daw, Arka and Meena, Muralikrishnan Gopalakrishnan and Lu, Hao}, journal = {AI4Science Workshop at International Conference on Machine Learning (ICML) 2024}, year = {2024}, publisher = {ICML}, category = {Workshop Papers} }
2023
- CVPR 2023
 Detecting and Tracking Hard-to-Detect Bacteria in Dense Porous BackgroundsMedha Sawhney, Bhas Karmarkar, Eric J Leaman, and 3 more authorsIn CVPR Workshop on CV4Animals 2023, 2023
Detecting and Tracking Hard-to-Detect Bacteria in Dense Porous BackgroundsMedha Sawhney, Bhas Karmarkar, Eric J Leaman, and 3 more authorsIn CVPR Workshop on CV4Animals 2023, 2023Studying bacteria motility is crucial to understanding and controlling biomedical and ecological phenomena involving bacteria. Tracking bacteria in complex environments such as polysaccharides (agar) or protein (collagen) hydrogels is a challenging task due to the lack of visually distinguishable features between bacteria and surrounding environment, making state-of-the-art methods for tracking easily recognizable objects such as pedestrians and cars unsuitable for this application. We propose a novel pipeline for detecting and tracking bacteria in bright-field microscopy videos involving bacteria in complex backgrounds. Our pipeline uses motion-based features and combines multiple models for detecting bacteria of varying difficulty levels. We apply multiple filters to prune false positive detections, and then use the SORT tracking algorithm with interpolation in case of missing detections. Our results demonstrate that our pipeline can accurately track hard-to-detect bacteria, achieving a high precision and recall.
@inproceedings{sawhney2023detecting, title = {Detecting and Tracking Hard-to-Detect Bacteria in Dense Porous Backgrounds}, author = {Sawhney, Medha and Karmarkar, Bhas and Leaman, Eric J and Daw, Arka and Karpatne, Anuj and Behkam, Bahareh}, year = {2023}, booktitle = {CVPR Workshop on CV4Animals 2023}, publisher = {CVPR}, category = {Workshop Papers} }
2022
- NeurIPS 2022
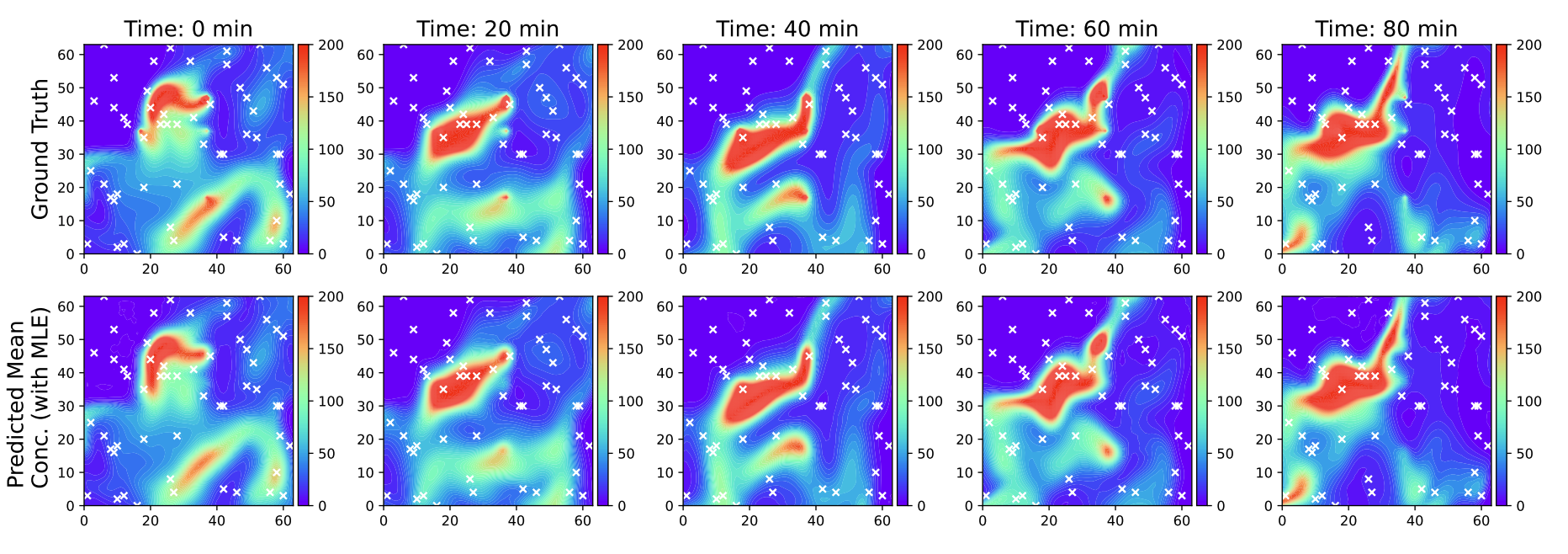 Source Identification and Field Reconstruction of Advection-Diffusion Process from Sparse Sensor MeasurementsArka Daw, Anuj Karpatne, Kyongmin Yeo, and 1 more authorIn NeurIPS Workshop on Machine Learning for Physical Sciences (ML4PS), 2022
Source Identification and Field Reconstruction of Advection-Diffusion Process from Sparse Sensor MeasurementsArka Daw, Anuj Karpatne, Kyongmin Yeo, and 1 more authorIn NeurIPS Workshop on Machine Learning for Physical Sciences (ML4PS), 2022Inferring the source information of greenhouse gases, such as methane, from spatially sparse sensor observations is an essential element in mitigating climate change. While it is well understood that the complex behavior of the atmospheric dispersion of such pollutants is governed by the Advection-Diffusion equation, it is difficult to directly apply the governing equations to identify the source information because of the spatially sparse observations, i.e., the pollution concentration is known only at the sensor locations. Here, we develop a multi-task learning framework that can provide high-fidelity reconstruction of the concentration field and identify emission characteristics of the pollution sources such as their location, emission strength, etc. from sparse sensor observations. We demonstrate that our proposed framework is able to achieve accurate reconstruction of the methane concentrations from sparse sensor measurements as well as precisely pin-point the location and emission strength of these pollution sources.
@inproceedings{daw2022source, title = {Source Identification and Field Reconstruction of Advection-Diffusion Process from Sparse Sensor Measurements}, author = {Daw, Arka and Karpatne, Anuj and Yeo, Kyongmin and Klein, Levente}, year = {2022}, booktitle = {NeurIPS Workshop on Machine Learning for Physical Sciences (ML4PS)}, publisher = {NeurIPS}, category = {Workshop Papers} }
2020
- NeurIPS 2020
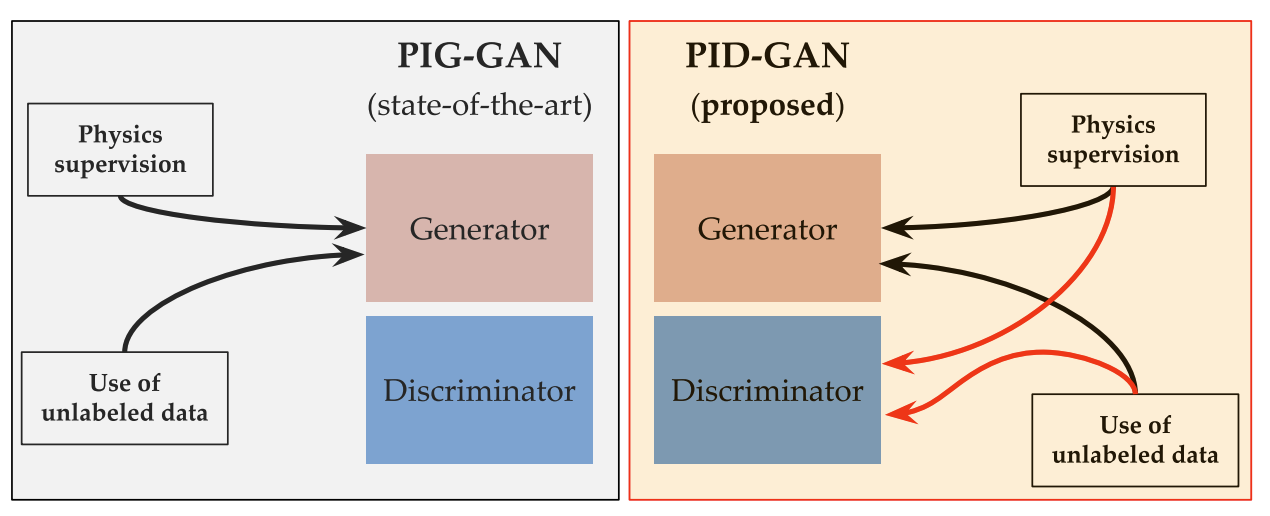 Physics-informed discriminator (PID) for conditional generative adversarial netsArka Daw, M Maruf, and Anuj KarpatneNeurIPS Workshop on Machine Learning for Physical Sciences (ML4PS), 2020
Physics-informed discriminator (PID) for conditional generative adversarial netsArka Daw, M Maruf, and Anuj KarpatneNeurIPS Workshop on Machine Learning for Physical Sciences (ML4PS), 2020We propose a novel method of incorporating physical knowledge as an additional input to the discriminator of a conditional Generative Adversarial Net (cGAN). Our proposed approach, termed as Physics-informed Discriminator for cGAN (cGAN-PID), is more aligned to the adversarial learning idea of cGAN as opposed to existing methods on incorporating physical knowledge in GANs by adding physics based loss functions as additional terms in the optimization objective of GAN. We evaluate the performance of our model on two toy datasets and demonstrate that our proposed cGAN-PID can be used as an alternative to the existing techniques.
@article{daw2020physics, title = {Physics-informed discriminator (PID) for conditional generative adversarial nets}, author = {Daw, Arka and Maruf, M and Karpatne, Anuj}, year = {2020}, publisher = {NeurIPS}, journal = {NeurIPS Workshop on Machine Learning for Physical Sciences (ML4PS)}, category = {Workshop Papers} }
2019
- KDD 2019
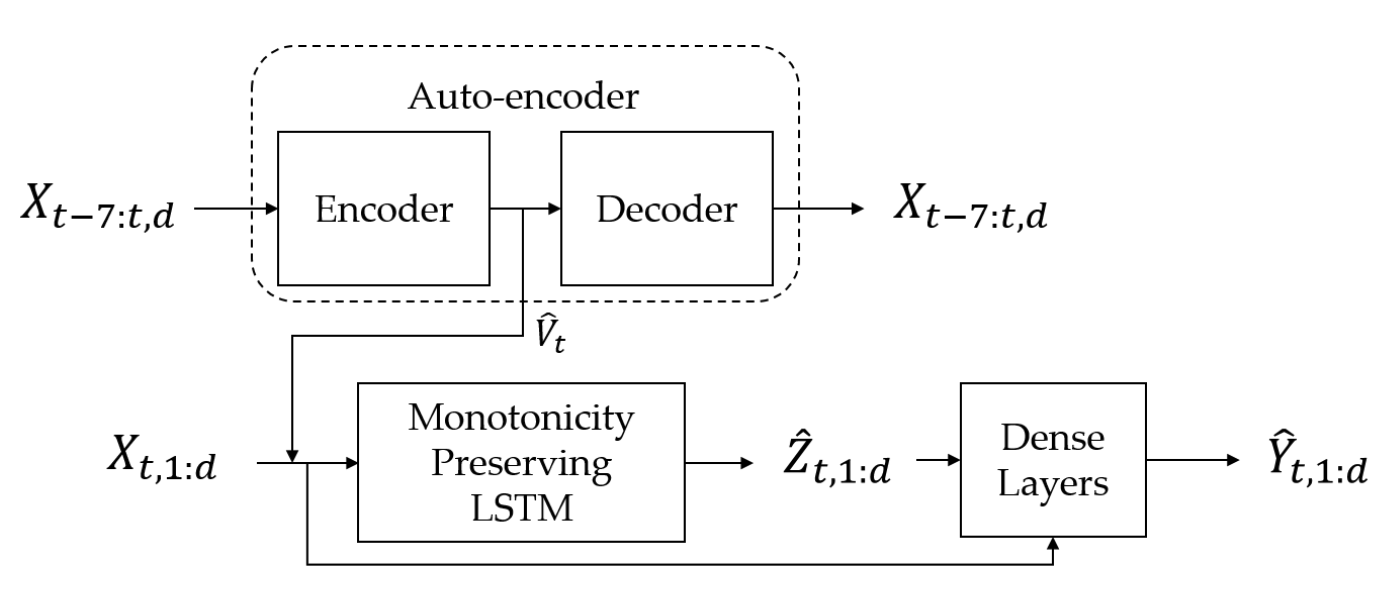 Physics-aware architecture of neural networks for uncertainty quantification: Application in lake temperature modelingArka Daw, and Anuj KarpatneIn , 2019
Physics-aware architecture of neural networks for uncertainty quantification: Application in lake temperature modelingArka Daw, and Anuj KarpatneIn , 2019In this paper, we explore a novel direction of research in theory-guided data science to develop physics-aware architectures of artificial neural networks (ANNs), where scientific knowledge is baked in the construction of ANN models. While previous efforts in theory-guided data science have used physics-based loss functions to guide the learning of neural network models to generalizable and physically consistent solutions, they do no guarantee that the model predictions will be physically consistent on unseen test instances, especially after slightly perturbing the trained model, as explored in dropout using testing methods for uncertainty quantification (UQ). On the other hand, our physics-aware ANN architecture hard-wires physical relationships in the ANN design, thus resulting in model predictions which always comply with known physics, even after performing dropout during testing for UQ. We provide some initial results to illustrate the effectiveness of our physics-aware neural network architecture in the context of lake temperature modeling, and show that our approach shows significantly lower physical inconsistency as compared to baseline methods.
@inproceedings{daw2019physics, title = {Physics-aware architecture of neural networks for uncertainty quantification: Application in lake temperature modeling}, author = {Daw, Arka and Karpatne, Anuj}, year = {2019}, organization = {SIGKDD}, journal = {Fragile Earth Workshop at Knowledge Discovery and Data Mining (KDD) 2019}, category = {Workshop Papers} }